Mass Spectral Filtering by Mass-Remainder Analysis (MARA) at High Resolution and Its Application to Metabolite Profiling of Flavonoids
Abstract
:1. Introduction
2. Results and Discussion
- (1)
- Calculation of the Mass-remainder values by Equation (1). MR3(O) and MR3(DBE) are calculated for all of the observed mass peaks (above an intensity threshold) of the ESI-DIMS mass spectrum. The divider base units (R1, R2, R3 in Equation (1)) are for M-MARA(O): 14.01565, 2.01565, and 0.0939, and for M-MARA(DBE): 15.994915, 1.97927, and 0.160795.
- (2)
- Rough filtering. The potential flavonoid MR3(O) and MR3(DBE) values are compiled in the MR-Table (see Figure 2b, Supplementary Table S1). A mass peak is removed, if its calculated MR3 values have not been found in the table.
- (3)
- Number of oxygen atom assignment and DBE value assignment. There is one-to-one correspondence between MR3(O) and the number of oxygen atoms in the molecule, and between MR3(DBE) and DBE value of the compound. The MR-Table (Table S1) can easily be used for these assignments.
- (4)
- Fine filtering. The correlations between the DBE versus m/z and number of oxygen atoms versus m/z values (see Figures S1 and S2) offer additional filtering criteria. Based on the flavonoid database [28,29], an m/z range was determined for every number of oxygen atoms and DBE value. If the m/z value of a mass peak does not fall into the ranges corresponding to its O and DBE values, the peak is eliminated.
- (5)
- Elemental composition determination. Once the number of oxygen atoms (O) and the DBE values have been assigned, the general formula of the compound follows as CxH2x+2-2*DBEOO, where x is the only unknown variable. Knowing the exact m/z of the deprotonated [M-H+]– ion, x can easily be calculated by Equation (2):where 1.007825, 1.007276, 15.994915, and 14.01565 are the exact masses of H, H+, oxygen and CH2, respectively.x = (m/z + 1.007276 − O × 15.994915 + 2 × DBE × 1.007825 − 2 × 1.007825)/14.01565
- (1)
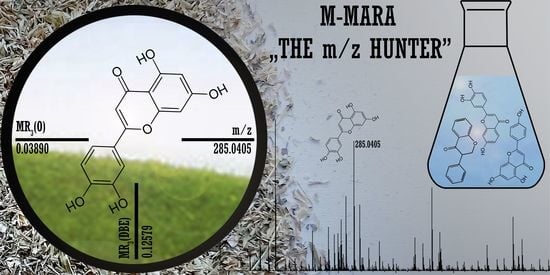
- MR3(O) = 0.0389, MR3(DBE) = 0.1258
- (2)
- The search in the MR-Table have found the calculated MR3 values, the compound is a potential flavonoid.
- (3)
- The number of oxygen atom and DBE values are O = 6, DBE = 11.
- (4)
- The m/z ranges corresponding to O = 6, DBE = 11 are [283.0236; 557.2921] and [221.0596; 655.2256], respectively. The measured m/z = 285.0405 falls into these intervals, the compound is accepted as a flavonoid.
- (5)
- Equation (2) gives x = 15, so the elemental composition of the compound is C15H10O6 (see Figure 4c)
3. Materials and Methods
3.1. Chemicals
3.2. Synthesis of Deuterium Labeled Rutin and Quercetin
3.3. Extraction of Samples
3.4. Electrospray Quadrupole Time-of-Flight Mass Spectrometry (ESI-QTOF MS)
3.5. Liquid Chromatography (HPLC-UV-MS)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.; Kopka, J.; Moritz, T. Plant metabolomics coming of age. Physiol. Plant. 2008, 132, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Markham, K.R. Flavonoids: Chemistry, Biochemistry and Applications; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Buckingham, J.; Munasinghe, V.R.N. Dictionary of Flavonoids with CD-ROM; Taylor & Francis: Abingdon, UK, 2015. [Google Scholar]
- Grotewold, E. The Science of Flavonoids; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Akimoto, N.; Ara, T.; Nakajima, D.; Suda, K.; Ikeda, C.; Takahashi, S.; Muneto, R.; Yamada, M.; Suzuki, H.; Shibata, D.; et al. FlavonoidSearch: A system for comprehensive flavonoid annotation by mass spectrometry. Sci. Rep. 2017, 7, 1243. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural Characterization of Flavonoid Glycoconjugates and Their Derivatives with Mass Spectrometric Techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Shi, L.; Zhang, F.; Chang, J.; Chu, X. High-throughput mass spectrometry scheme for screening and quantification of flavonoids in antioxidant nutraceuticals. J. Chromatogr. A 2019, 1608, 460408. [Google Scholar] [CrossRef]
- Bingol, K.; Bruschweiler-Li, L.; Yu, C.; Somogyi, A.; Zhang, F.; Brüschweiler, R. Metabolomics Beyond Spectroscopic Databases: A Combined MS/NMR Strategy for the Rapid Identification of New Metabolites in Complex Mixtures. Anal. Chem. 2015, 87, 3864–3870. [Google Scholar] [CrossRef] [Green Version]
- Timári, I.; Wang, C.; Hansen, A.L.; Costa dos Santos, G.; Yoon, S.O.; Bruschweiler-Li, L.; Brüschweiler, R. Real-Time Pure Shift HSQC NMR for Untargeted Metabolomics. Anal. Chem. 2019, 91, 2304–2311. [Google Scholar] [CrossRef]
- Fuhrer, T.; Heer, D.; Begemann, B.; Zamboni, N. High-Throughput, Accurate Mass Metabolome Profiling of Cellular Extracts by Flow Injection–Time-of-Flight Mass Spectrometry. Anal. Chem. 2011, 83, 7074–7080. [Google Scholar] [CrossRef]
- Sleno, L. The use of mass defect in modern mass spectrometry. J. Mass Spectrom. 2012, 47, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Ma, L.; Zhang, D.; Ray, K.; Zhao, W.; Humphreys, W.G.; Skiles, G.; Sanders, M.; Zhang, H. Detection and Characterization of Metabolites in Biological Matrices Using Mass Defect Filtering of Liquid Chromatography/High Resolution Mass Spectrometry Data. Drug Metab. Dispos. 2006, 34, 1722–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, D.; Ray, K.; Zhu, M. Mass defect filter technique and its applications to drug metabolite identification by high-resolution mass spectrometry. J. Mass Spectrom. 2009, 44, 999–1016. [Google Scholar] [CrossRef] [PubMed]
- Ekanayaka, E.A.P.; Celiz, M.D.; Jones, A.D. Relative Mass Defect Filtering of Mass Spectra: A Path to Discovery of Plant Specialized Metabolites. Plant Physiol. 2015, 167, 1221–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Rodgers, R.P.; Marshall, A.G. Truly “exact” mass: Elemental composition can be determined uniquely from molecular mass measurement at ∼0.1 mDa accuracy for molecules up to ∼500 Da. Int. J. Mass Spectrom. 2006, 251, 260–265. [Google Scholar] [CrossRef]
- Kendrick, E. A Mass Scale Based on CH2 = 14.0000 for High Resolution Mass Spectrometry of Organic Compounds. Anal. Chem. 1963, 35, 2146–2154. [Google Scholar] [CrossRef]
- Fouquet, T.; Satoh, T.; Sato, H. First Gut Instincts Are Always Right: The Resolution Required for a Mass Defect Analysis of Polymer Ions Can Be as Low as Oligomeric. Anal. Chem. 2018, 90, 2404–2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouquet, T.N.J. The Kendrick analysis for polymer mass spectrometry. J. Mass Spectrom. 2019, 54, 933–947. [Google Scholar] [CrossRef]
- Fouquet, T.N.J.; Pizzala, H.; Rollet, M.; Crozet, D.; Giusti, P.; Charles, L. Mass Spectrometry-Based Analytical Strategy for Comprehensive Molecular Characterization of Biodegradable Poly(lactic-co-glycolic Acid) Copolymers. J. Am. Soc. Mass. Spectrom. 2020, 31, 1554–1562. [Google Scholar] [CrossRef]
- Nagy, T.; Kuki, Á.; Zsuga, M.; Kéki, S. Mass-Remainder Analysis (MARA): A New Data Mining Tool for Copolymer Characterization. Anal. Chem. 2018, 90, 3892–3897. [Google Scholar] [CrossRef]
- Nagy, T.; Kuki, Á.; Nagy, M.; Zsuga, M.; Kéki, S. Mass-Remainder Analysis (MARA): An Improved Method for Elemental Composition Assignment in Petroleomics. Anal. Chem. 2019, 91, 6479–6486. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.; Kuki, Á.; Hashimov, M.; Zsuga, M.; Kéki, S. Multistep Mass-Remainder Analysis and its Application in Copolymer Blends. Macromolecules 2020, 53, 1199–1204. [Google Scholar] [CrossRef]
- Hashimov, M.; Kuki, Á.; Nagy, T.; Zsuga, M.; Kéki, S. Tandem Mass-Remainder Analysis of Industrially Important Polyether Polyols. Polymers 2020, 12, 2768. [Google Scholar] [CrossRef] [PubMed]
- Kuki, Á.; Nagy, T.; Hashimov, M.; File, S.; Nagy, M.; Zsuga, M.; Kéki, S. Mass Spectrometric Characterization of Epoxidized Vegetable Oils. Polymers 2019, 11, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flavonoid Database, Arita Laboratory, National Institute of Genetics. Available online: http://metabolomics.jp/wiki/Main_Page (accessed on 5 October 2020).
- Arita, M.; Suwa, K. Search extension transforms Wiki into a relational system: A case for flavonoid metabolite database. BioData Mining 2008, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.-H.; Li, Y.; Zhang, M.-L.; Wang, Y.-F.; Zhang, Q.; Qin, F.; Shi, Q.-W.; Kiyota, H. Cytotoxic flavonoids from the flowers of Achillea millefolium. Chem. Nat. Compd. 2013, 48, 958–962. [Google Scholar] [CrossRef]
- Kruve, A. Strategies for Drawing Quantitative Conclusions from Nontargeted Liquid Chromatography–High-Resolution Mass Spectrometry Analysis. Anal. Chem. 2020, 92, 4691–4699. [Google Scholar] [CrossRef] [Green Version]
- Hiraoka, K.; Miyamoto, T.; Baba, S.; Furuta, T. Preparation of deuterium-labeled rutin by hydrogen exchange reaction. J. Label. Compd. Radiopharm. 1981, 18, 613–619. [Google Scholar] [CrossRef]







| Type | Yarrow (Achillea millefolum L.) | Elder (Sambucus nigra L.) | Birch (Betula L.) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| No. of peaks | in the spectra | 1258 | 889 | 883 | 1254 | 842 | 1070 | 1516 | 1251 | 1293 | 1067 | 1033 | 1578 | 1180 | 1662 | 1590 |
| M-MARA | 66 | 15 | 52 | 65 | 53 | 58 | 56 | 49 | 35 | 52 | 40 | 60 | 57 | 67 | 63 | |
| database search | 51 | 9 | 36 | 45 | 40 | 45 | 43 | 37 | 28 | 41 | 31 | 41 | 45 | 45 | 46 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagy, T.; Róth, G.; Kuki, Á.; Zsuga, M.; Kéki, S. Mass Spectral Filtering by Mass-Remainder Analysis (MARA) at High Resolution and Its Application to Metabolite Profiling of Flavonoids. Int. J. Mol. Sci. 2021, 22, 864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020864
Nagy T, Róth G, Kuki Á, Zsuga M, Kéki S. Mass Spectral Filtering by Mass-Remainder Analysis (MARA) at High Resolution and Its Application to Metabolite Profiling of Flavonoids. International Journal of Molecular Sciences. 2021; 22(2):864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020864
Chicago/Turabian StyleNagy, Tibor, Gergő Róth, Ákos Kuki, Miklós Zsuga, and Sándor Kéki. 2021. "Mass Spectral Filtering by Mass-Remainder Analysis (MARA) at High Resolution and Its Application to Metabolite Profiling of Flavonoids" International Journal of Molecular Sciences 22, no. 2: 864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020864