Recruitment of M1 Macrophages May Not Be Critical for Protection against Colitis-Associated Tumorigenesis
Abstract
:1. Introduction
2. Results
2.1. The Complete Absence of the IL-4 Receptor α-Chain Inhibits Colon Tumor Development
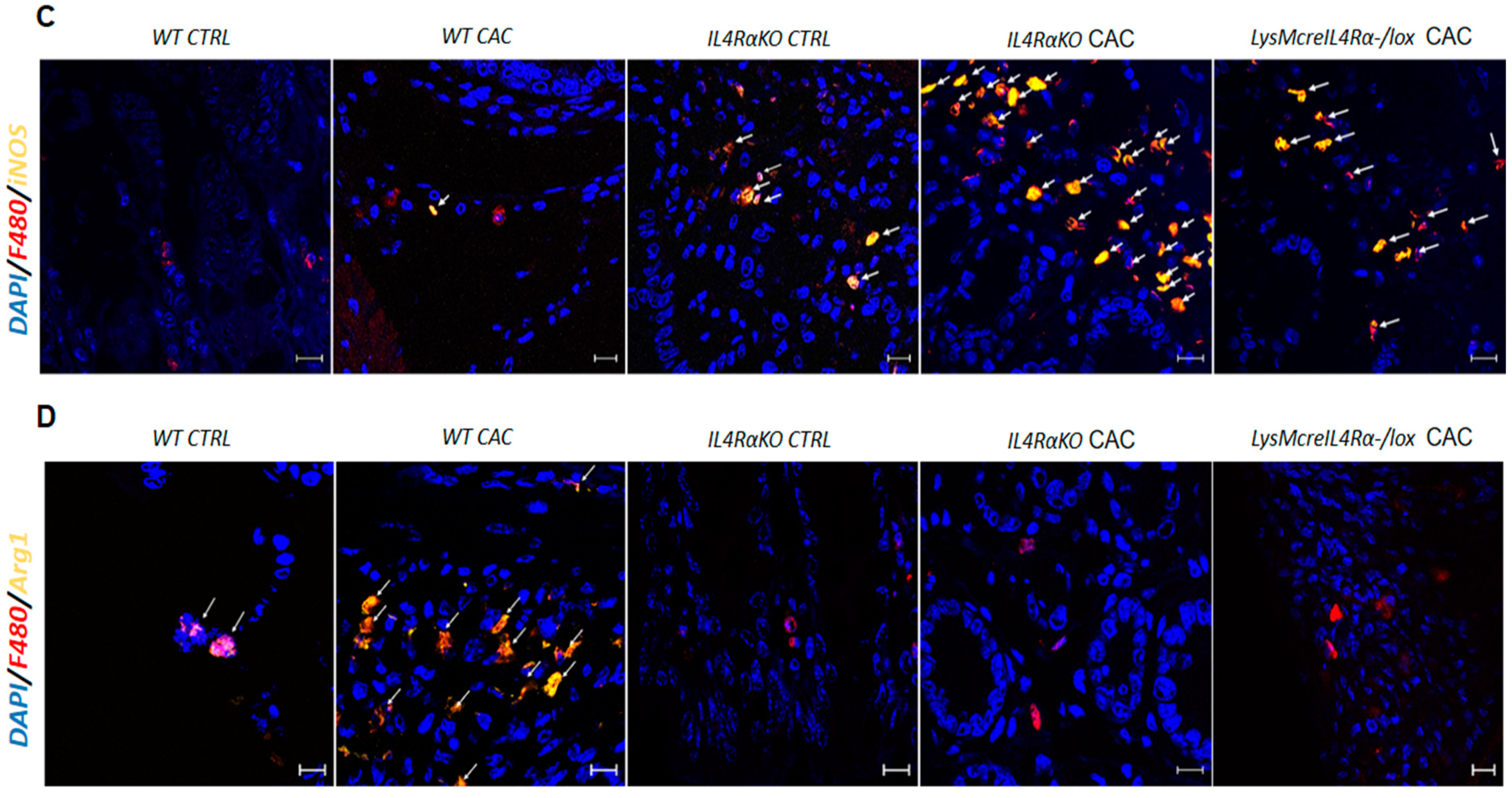
2.2. The Absence of IL4Rα Favors M1 Macrophage Recruitment in the Colon
2.3. IL4Rα Expression in Macrophages Is Not Directly Related to Tumor Development in CAC
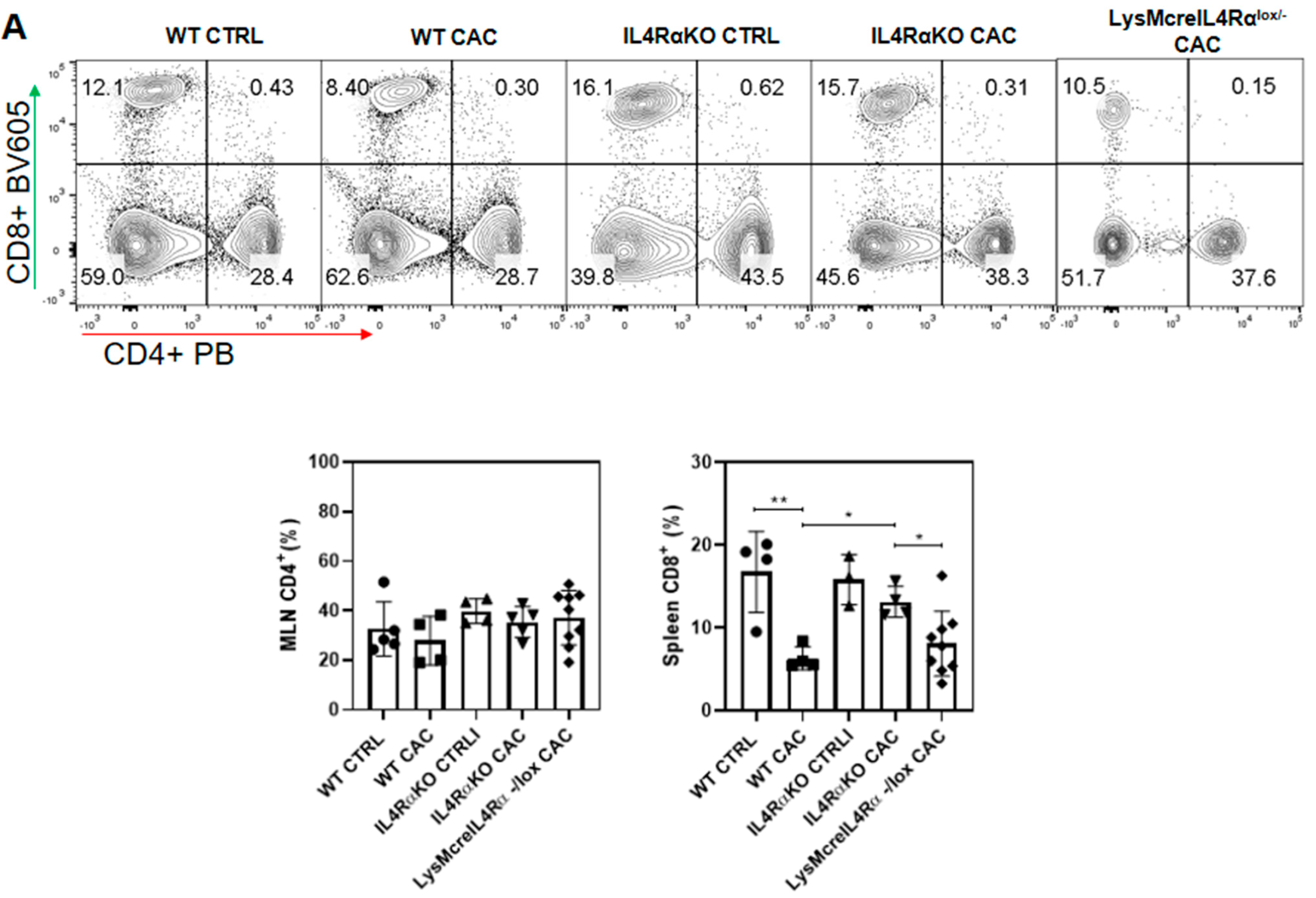
2.4. Colon Infiltration of M1 Macrophages in LysMcreIL4Rα−/lox-CAC Mice Is Not Enough to Avoid Colon Tumorigenesis
2.5. M1 Macrophages in LysMcreIL4Rα−/lox Mice Do Not Have a Protective Role during CAC
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Generation of LysMcreIL-4Rα−/lox BALB/c Mice
4.3. Induction of Colitis Associated Colon Cancer (CAC) Model
4.4. Histological Analysis
4.5. Immunofluorescence (IF)
4.6. Isolation of Cells from Colon Tumors for Flow Cytometry
4.7. Flow Cytometry
4.8. Transcription Factors in Colon Cells for Flow Cytometry
4.9. Cell Culture and Cytokine Quantification
4.10. RNA Isolation and RT–PCR
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Dyson, J.K.; Rutter, M.D. Colorectal cancer in inflammatory bowel disease: What is the real magnitude of the risk? World J. Gastroenterol. 2012, 18, 3839–3848. [Google Scholar] [CrossRef]
- Terzic, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e2105. [Google Scholar] [CrossRef] [PubMed]
- Ingram, N.; Northwood, E.L.; Perry, S.L.; Marston, G.; Snowden, H.; Taylor, J.C.; Scott, N.; Bishop, D.T.; Coletta, P.L.; Hull, M.A. Reduced type II interleukin-4 receptor signalling drives initiation, but not progression, of colorectal carcinogenesis: Evidence from transgenic mouse models and human case-control epidemiological observations. Carcinogenesis 2013, 34, 2341–2349. [Google Scholar] [CrossRef] [PubMed]
- Koller, F.L.; Hwang, D.G.; Dozier, E.A.; Fingleton, B. Epithelial interleukin-4 receptor expression promotes colon tumor growth. Carcinogenesis 2010, 31, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Bankaitis, K.V.; Fingleton, B. Targeting IL4/IL4R for the treatment of epithelial cancer metastasis. Clin. Exp. Metastasis 2015, 32, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Vinnakota, K.; Zhang, Y.; Selvanesan, B.C.; Topi, G.; Salim, T.; Sand-Dejmek, J.; Jonsson, G.; Sjolander, A. M2-like macrophages induce colon cancer cell invasion via matrix metalloproteinases. J. Cell. Physiol. 2017, 232, 3468–3480. [Google Scholar] [CrossRef]
- Wang, H.W.; Joyce, J.A. Alternative activation of tumor-associated macrophages by IL-4: Priming for protumoral functions. Cell Cycle 2010, 9, 4824–4835. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Chen, B.; Yang, Z. The Role of Tumor-Associated Macrophages in Colorectal Carcinoma Progression. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 45, 356–365. [Google Scholar] [CrossRef]
- Duluc, D.; Corvaisier, M.; Blanchard, S.; Catala, L.; Descamps, P.; Gamelin, E.; Ponsoda, S.; Delneste, Y.; Hebbar, M.; Jeannin, P. Interferon-gamma reverses the immunosuppressive and protumoral properties and prevents the generation of human tumor-associated macrophages. Int. J. Cancer 2009, 125, 367–373. [Google Scholar] [CrossRef]
- Seril, D.N.; Liao, J.; Yang, G.Y. Colorectal carcinoma development in inducible nitric oxide synthase-deficient mice with dextran sulfate sodium-induced ulcerative colitis. Mol. Carcinog. 2007, 46, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Ma, A.; Urbanski, S.J.; McCafferty, D.M. Induction of inducible nitric oxide synthase: A protective mechanism in colitis-induced adenocarcinoma. Carcinogenesis 2007, 28, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Adams, C.; McCarthy, H.O.; Coulter, J.A.; Worthington, J.; Murphy, C.; Robson, T.; Hirst, D.G. Nitric oxide synthase gene therapy enhances the toxicity of cisplatin in cancer cells. J. Gene Med. 2009, 11, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Saito, K.; Shiiba, K.; Ohuchi, A.; Saigenji, K.; Nagura, H.; Ohtani, H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998, 58, 3491–3494. [Google Scholar] [PubMed]
- Pozzi, L.A.; Maciaszek, J.W.; Rock, K.L. Both dendritic cells and macrophages can stimulate naive CD8 T cells in vivo to proliferate, develop effector function, and differentiate into memory cells. J. Immunol. 2005, 175, 2071–2081. [Google Scholar] [CrossRef]
- Ong, S.M.; Tan, Y.C.; Beretta, O.; Jiang, D.; Yeap, W.H.; Tai, J.J.; Wong, W.C.; Yang, H.; Schwarz, H.; Lim, K.H.; et al. Macrophages in human colorectal cancer are pro-inflammatory and prime T cells towards an anti-tumour type-1 inflammatory response. Eur. J. Immunol. 2012, 42, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Cui, Y.L.; Li, H.K.; Zhou, H.Y.; Zhang, T.; Li, Q. Correlations of tumor-associated macrophage subtypes with liver metastases of colorectal cancer. Asian Pac. J. Cancer Prev. 2013, 14, 1003–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.C.; Chen, J.S.; Lee, C.H.; Chang, J.J.; Shieh, Y.S. Intratumoral macrophage counts correlate with tumor progression in colorectal cancer. J. Surg. Oncol. 2010, 102, 242–248. [Google Scholar] [CrossRef]
- Bogen, B.; Fauskanger, M.; Haabeth, O.A.; Tveita, A. CD4(+) T cells indirectly kill tumor cells via induction of cytotoxic macrophages in mouse models. Cancer Immunol. Immunother. 2019, 68, 1865–1873. [Google Scholar] [CrossRef]
- Muller, E.; Christopoulos, P.F.; Halder, S.; Lunde, A.; Beraki, K.; Speth, M.; Oynebraten, I.; Corthay, A. Toll-Like Receptor Ligands and Interferon-gamma Synergize for Induction of Antitumor M1 Macrophages. Front. Immunol. 2017, 8, 1383. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, H.; Chen, B.; Liu, X.; Zhang, S.; Zong, Z.; Gao, M. PD-L1-Mediated Immunosuppression in Glioblastoma Is Associated With the Infiltration and M2-Polarization of Tumor-Associated Macrophages. Front. Immunol. 2020, 11, 588552. [Google Scholar] [CrossRef]
- Fauskanger, M.; Haabeth, O.A.W.; Skjeldal, F.M.; Bogen, B.; Tveita, A.A. Tumor Killing by CD4(+) T Cells Is Mediated via Induction of Inducible Nitric Oxide Synthase-Dependent Macrophage Cytotoxicity. Front. Immunol. 2018, 9, 1684. [Google Scholar] [CrossRef] [Green Version]
- Dinapoli, M.R.; Calderon, C.L.; Lopez, D.M. The altered tumoricidal capacity of macrophages isolated from tumor-bearing mice is related to reduce expression of the inducible nitric oxide synthase gene. J. Exp. Med. 1996, 183, 1323–1329. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, N.E. Macrophages and Dendritic Cells. Methods and Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; pp. v–vi. [Google Scholar] [CrossRef]
- Mehta, A.K.; Gracias, D.T.; Croft, M. TNF activity and T cells. Cytokine 2018, 101, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Leon-Cabrera, S.A.; Molina-Guzman, E.; Delgado-Ramirez, Y.G.; Vazquez-Sandoval, A.; Ledesma-Soto, Y.; Perez-Plasencia, C.G.; Chirino, Y.I.; Delgado-Buenrostro, N.L.; Rodriguez-Sosa, M.; Vaca-Paniagua, F.; et al. Lack of STAT6 Attenuates Inflammation and Drives Protection against Early Steps of Colitis-Associated Colon Cancer. Cancer Immunol. Res. 2017, 5, 385–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ries, C.H.; Cannarile, M.A.; Hoves, S.; Benz, J.; Wartha, K.; Runza, V.; Rey-Giraud, F.; Pradel, L.P.; Feuerhake, F.; Klaman, I.; et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 2014, 25, 846–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef]
- Keller, R.; Geiges, M.; Keist, R. L-arginine-dependent reactive nitrogen intermediates as mediators of tumor cell killing by activated macrophages. Cancer Res. 1990, 50, 1421–1425. [Google Scholar]
- Cui, S.; Reichner, J.S.; Mateo, R.B.; Albina, J.E. Activated murine macrophages induce apoptosis in tumor cells through nitric oxide-dependent or -independent mechanisms. Cancer Res. 1994, 54, 2462–2467. [Google Scholar]
- Vicetti Miguel, R.D.; Cherpes, T.L.; Watson, L.J.; McKenna, K.C. CTL induction of tumoricidal nitric oxide production by intratumoral macrophages is critical for tumor elimination. J. Immunol. 2010, 185, 6706–6718. [Google Scholar] [CrossRef] [Green Version]
- Albina, J.E.; Reichner, J.S. Role of nitric oxide in mediation of macrophage cytotoxicity and apoptosis. Cancer Metastasis Rev. 1998, 17, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Huang, S.; Dong, Z.; Juang, S.H.; Gutman, M.; Xie, Q.W.; Nathan, C.; Fidler, I.J. Transfection with the inducible nitric oxide synthase gene suppresses tumorigenicity and abrogates metastasis by K-1735 murine melanoma cells. J. Exp. Med. 1995, 181, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Old, L.J.; Schreiber, R.D. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002, 13, 95–109. [Google Scholar] [CrossRef]
- Corthay, A.; Skovseth, D.K.; Lundin, K.U.; Rosjo, E.; Omholt, H.; Hofgaard, P.O.; Haraldsen, G.; Bogen, B. Primary antitumor immune response mediated by CD4+ T cells. Immunity 2005, 22, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, T.; Ramakrishnan, M.; Diamanti, M.A.; Ziegler, P.K.; Brombacher, F.; Greten, F.R. Loss of Stat6 affects chromatin condensation in intestinal epithelial cells causing diverse outcome in murine models of inflammation-associated and sporadic colon carcinogenesis. Oncogene 2019, 38, 1787–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fichtner-Feigl, S.; Strober, W.; Kawakami, K.; Puri, R.K.; Kitani, A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat. Med. 2006, 12, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, R.; Kawamura, Y.I.; Kato, R.; Mizutani, N.; Toyama-Sorimachi, N.; Dohi, T. IL-13 receptor alpha2 promotes epithelial cell regeneration from radiation-induced small intestinal injury in mice. Gastroenterology 2006, 131, 130–141. [Google Scholar] [CrossRef]
- Aliberti, J.; Hieny, S.; Reis e Sousa, C.; Serhan, C.N.; Sher, A. Lipoxin-mediated inhibition of IL-12 production by DCs: A mechanism for regulation of microbial immunity. Nat. Immunol. 2002, 3, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Song, Z.; Chu, J.; Qu, X. Deficiency of interferon-gamma or its receptor promotes colorectal cancer development. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2015, 35, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Luissint, A.C.; Parkos, C.A.; Nusrat, A. Inflammation and the Intestinal Barrier: Leukocyte-Epithelial Cell Interactions, Cell Junction Remodeling, and Mucosal Repair. Gastroenterology 2016, 151, 616–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, E.M.; Ryu, S.H.; Singh, A.P.; Lee, G.; Goretsky, T.; Sinh, P.; Williams, D.B.; Cloud, A.L.; Gounaris, E.; Patel, V.; et al. Epithelial TNF Receptor Signaling Promotes Mucosal Repair in Inflammatory Bowel Disease. J. Immunol. 2017, 199, 1886–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Han, G.; Chen, Y.; Wang, K.; Liu, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Shen, B.; et al. Protective role of tumor necrosis factor (TNF) receptors in chronic intestinal inflammation: TNFR1 ablation boosts systemic inflammatory response. Lab. Investig. A J. Tech. Methods Pathol. 2013, 93, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Hale, L.P.; Greer, P.K. A novel murine model of inflammatory bowel disease and inflammation-associated colon cancer with ulcerative colitis-like features. PLoS ONE 2012, 7, e41797. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-gamma in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Klampfer, L. Cytokines, inflammation and colon cancer. Curr. Cancer Drug Targets 2011, 11, 451–464. [Google Scholar] [CrossRef]
- Mager, L.F.; Wasmer, M.H.; Rau, T.T.; Krebs, P. Cytokine-Induced Modulation of Colorectal Cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef]
- Roth, F.; De La Fuente, A.C.; Vella, J.L.; Zoso, A.; Inverardi, L.; Serafini, P. Aptamer-mediated blockade of IL4Ralpha triggers apoptosis of MDSCs and limits tumor progression. Cancer Res. 2012, 72, 1373–1383. [Google Scholar] [CrossRef] [Green Version]
- Gnant, M.; Mlineritsch, B.; Stoeger, H.; Luschin-Ebengreuth, G.; Heck, D.; Menzel, C.; Jakesz, R.; Seifert, M.; Hubalek, M.; Pristauz, G.; et al. Adjuvant endocrine therapy plus zoledronic acid in premenopausal women with early-stage breast cancer: 62-month follow-up from the ABCSG-12 randomised trial. Lancet. Oncol. 2011, 12, 631–641. [Google Scholar] [CrossRef]
- Kim, E.Y.; Teh, H.S. TNF type 2 receptor (p75) lowers the threshold of T cell activation. J. Immunol. 2001, 167, 6812–6820. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.Y.; Priatel, J.J.; Teh, S.J.; Teh, H.S. TNF receptor type 2 (p75) functions as a costimulator for antigen-driven T cell responses in vivo. J. Immunol. 2006, 176, 1026–1035. [Google Scholar] [CrossRef] [Green Version]
- Schubart, C.; Krljanac, B.; Otte, M.; Symowski, C.; Martini, E.; Gunther, C.; Becker, C.; Daniel, C.; Voehringer, D. Selective expression of constitutively activated STAT6 in intestinal epithelial cells promotes differentiation of secretory cells and protection against helminths. Mucosal Immunol. 2019, 12, 413–424. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Li, B.; Yang, X.; Liu, T.; Shi, T.; Deng, B.; Zhang, Y.; Jia, L.; Jiang, Z.; He, R. Non-hematopoietic STAT6 induces epithelial tight junction dysfunction and promotes intestinal inflammation and tumorigenesis. Mucosal Immunol. 2019, 12, 1304–1315. [Google Scholar] [CrossRef]
- Fukushi, J.; Ono, M.; Morikawa, W.; Iwamoto, Y.; Kuwano, M. The activity of soluble VCAM-1 in angiogenesis stimulated by IL-4 and IL-13. J. Immunol. 2000, 165, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Fukushi, J.; Morisaki, T.; Shono, T.; Nishie, A.; Torisu, H.; Ono, M.; Kuwano, M. Novel biological functions of interleukin-4: Formation of tube-like structures by vascular endothelial cells in vitro and angiogenesis in vivo. Biochem. Biophys. Res. Commun. 1998, 250, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Conticello, C.; Pedini, F.; Zeuner, A.; Patti, M.; Zerilli, M.; Stassi, G.; Messina, A.; Peschle, C.; De Maria, R. IL-4 protects tumor cells from anti-CD95 and chemotherapeutic agents via up-regulation of antiapoptotic proteins. J. Immunol. 2004, 172, 5467–5477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, M.; Kobayashi, H.; Pollard, R.B.; Suzuki, F. A pathogenic role of Th2 cells and their cytokine products on the pulmonary metastasis of murine B16 melanoma. J. Immunol. 1998, 160, 5869–5873. [Google Scholar] [PubMed]
- Dufort, F.J.; Bleiman, B.F.; Gumina, M.R.; Blair, D.; Wagner, D.J.; Roberts, M.F.; Abu-Amer, Y.; Chiles, T.C. Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. J. Immunol. 2007, 179, 4953–4957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venmar, K.T.; Kimmel, D.W.; Cliffel, D.E.; Fingleton, B. IL4 receptor alpha mediates enhanced glucose and glutamine metabolism to support breast cancer growth. Biochim. Biophys. Acta 2015, 1853, 1219–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbert, D.R.; Holscher, C.; Mohrs, M.; Arendse, B.; Schwegmann, A.; Radwanska, M.; Leeto, M.; Kirsch, R.; Hall, P.; Mossmann, H.; et al. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 2004, 20, 623–635. [Google Scholar] [CrossRef]
- Brombacher, F.; Arendse, B.; Peterson, R.; Holscher, A.; Holscher, C. Analyzing classical and alternative macrophage activation in macrophage/neutrophil-specific IL-4 receptor-alpha-deficient mice. Methods Mol. Biol. 2009, 531, 225–252. [Google Scholar] [CrossRef]
- Tanaka, T.; Kohno, H.; Suzuki, R.; Yamada, Y.; Sugie, S.; Mori, H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003, 94, 965–973. [Google Scholar] [CrossRef]








| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| IL4Rα Wild Type | TGACCTACAAGGAACCCAGGC | CTCGGCGCACTGACCCATCT |
| IL4Rα Deleted | GGCTGCTGACCTGGAATAACC | CCTTTGAGAACTGCGGGCT |
| IL4Rα lox | CCCTTCCTGGCCCTGAATTT | GTTTCCTCCTACCGCTGATT |
| LysMcre | CTTGGGCTGCCAGAATTTCTC | CCCAGAAATGCCAGATTACG |
| IL13Rα2 | ATA CGT ACG CAT TTG TCA GAG CA | CCA AGC CCT CAT ACC AGA AAA AC |
| Arginase 1 | CAGAAGAATGGAAGAGTCAG | CAGATATGCAGGGAGTCACC |
| Relmα | GGTCCCAGTGCATATGGATGAGACCATAGA | CACCTCTTCACTCGAGGGACAGTTGGCAGC |
| iNOS | CTGGAGGAGCTCCTGCCTCATG | GCAGCATCCCCTCTGATGGTG |
| GAPDH | CTCATGACCACAGTCCATGC | CACATTGGGGGTAGGAACAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Andrade, I.; Olguín, J.E.; Guerrero-García, S.; Espinosa, J.A.; Garduño-Javier, E.; Hernández-Gómez, V.; Vaca-Paniagua, F.; Rodríguez-Sosa, M.; Terrazas, L.I. Recruitment of M1 Macrophages May Not Be Critical for Protection against Colitis-Associated Tumorigenesis. Int. J. Mol. Sci. 2021, 22, 11204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222011204
Medina-Andrade I, Olguín JE, Guerrero-García S, Espinosa JA, Garduño-Javier E, Hernández-Gómez V, Vaca-Paniagua F, Rodríguez-Sosa M, Terrazas LI. Recruitment of M1 Macrophages May Not Be Critical for Protection against Colitis-Associated Tumorigenesis. International Journal of Molecular Sciences. 2021; 22(20):11204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222011204
Chicago/Turabian StyleMedina-Andrade, Itzel, Jonadab E. Olguín, Stephanie Guerrero-García, Jossael A. Espinosa, Elizabeth Garduño-Javier, Victoria Hernández-Gómez, Felipe Vaca-Paniagua, Miriam Rodríguez-Sosa, and Luis I. Terrazas. 2021. "Recruitment of M1 Macrophages May Not Be Critical for Protection against Colitis-Associated Tumorigenesis" International Journal of Molecular Sciences 22, no. 20: 11204. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222011204








