Neochlorogenic Acid Attenuates Hepatic Lipid Accumulation and Inflammation via Regulating miR-34a In Vitro
Abstract
:1. Introduction
2. Results
2.1. Effect of 5-CQA and OA on the Viability of HepG2 Cells
2.2. Effect of 5-CQA on OA-Induced Lipid Accumulation
2.3. Effect of 5-CQA on OA-Induced Fatty Acid and Cholesterol Synthesis
2.4. Effect of 5-CQA on OA-Induced Defect of Fatty Acid Oxidation and Inflammation
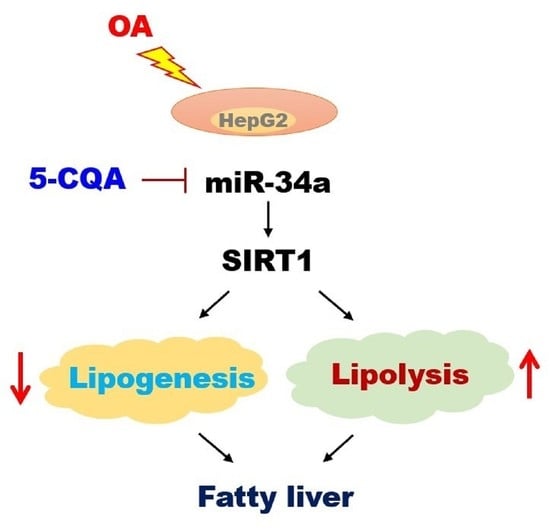
2.5. Effect of 5-CQA on the miR-34a/SIRT1/AMPK Pathway
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. MTT Assay
4.4. Oil Red O Staining
4.5. Nile Red Flow Cytometry Analysis
4.6. miRNA Extraction and Real-Time PCR
4.7. Transfection with miR-34a Mimic and Inhibitor
4.8. Western Blot Analysis
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaggini, M.; Morelli, M.; Buzzigoli, E.; DeFronzo, R.A.; Bugianesi, E.; Gastaldelli, A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients 2013, 5, 1544–1560. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Non-alcoholic fatty liver disease. BMC Med. 2017, 15, 45. [Google Scholar] [CrossRef] [Green Version]
- Serviddio, G.; Bellanti, F.; Vendemiale, G. Free radical biology for medicine: Learning from nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2013, 65, 952–968. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef]
- Horton, J.D.; Shah, N.A.; Warrington, J.A.; Anderson, N.N.; Park, S.W.; Brown, M.S.; Goldstein, J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 2003, 100, 12027–12032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, T.Y.; Lin, S.-m.; Leung, L.K. The flavone luteolin suppresses SREBP-2 expression and post-translational activation in hepatic cells. PLoS ONE 2015, 10, e0135637. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Gao, Y.; Zhang, L.; Yin, Y.; Zhang, W. Rspo1/Rspo3-LGR4 signaling inhibits hepatic cholesterol synthesis through the AMPKα-SREBP2 pathway. FASEB J. 2020, 34, 14946–14959. [Google Scholar] [CrossRef]
- Price, N.L.; Gomes, A.P.; Ling, A.J.; Duarte, F.V.; Martin-Montalvo, A.; North, B.J.; Agarwal, B.; Ye, L.; Ramadori, G.; Teodoro, J.S. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012, 15, 675–690. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Liu, S.; Zhai, A.; Zhang, B.; Tian, G. AMPK-mediated regulation of lipid metabolism by phosphorylation. Biol. Pharm. Bull. 2018, 41, 985–993. [Google Scholar] [CrossRef] [Green Version]
- Chyau, C.-C.; Wang, H.-F.; Zhang, W.-J.; Chen, C.-C.; Huang, S.-H.; Chang, C.-C.; Peng, R.Y. Antrodan alleviates high-fat and high-fructose diet-induced fatty liver disease in C57BL/6 mice model via AMPK/Sirt1/SREBP-1c/PPARγ pathway. Int. J. Mol. Sci. 2020, 21, 360. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yan, Y.; Xv, W.; Qian, G.; Li, C.; Zou, H.; Li, Y. A new insight into the roles of MiRNAs in metabolic syndrome. BioMed Res. Int. 2018, 2018, 7372636. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Mohamed, I.N.; Teoh, S.L.; Thevaraj, T.; Ku Ahmad Nasir, K.N.; Zawawi, A.; Salim, H.H.; Zhou, D.K. Micro-RNA and the features of metabolic syndrome: A narrative review. Mini Rev. Med. Chem. 2020, 20, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Biersack, B. Current state of phenolic and terpenoidal dietary factors and natural products as non-coding RNA/microRNA modulators for improved cancer therapy and prevention. Non-Coding RNA Res. 2016, 1, 12–34. [Google Scholar] [CrossRef] [Green Version]
- Zobeiri, M.; Parvizi, F.; Kalhori, M.R.; Majnooni, M.B.; Farzaei, M.H.; Abdollahi, M. Targeting miRNA by Natural Products: A Novel Therapeutic Approach for Nonalcoholic Fatty Liver. Evid.-Based Complementary Altern. Med. 2021, 2021, 6641031. [Google Scholar] [CrossRef]
- Torres, L.; Cogliati, B.; Otton, R. Green tea prevents NAFLD by modulation of miR-34a and miR-194 expression in a high-fat diet mouse model. Oxidative Med. Cell. Longev. 2019, 2019, 4168380. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhu, Y.; Hu, S.; Pan, X.; Bawa, F.C.; Wang, H.H.; Wang, D.Q.-H.; Yin, L.; Zhang, Y. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol. Metab. 2021, 51, 101244. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Hsu, J.-D.; Lin, W.-L.; Kao, S.-H.; Wang, C.-J. Upregulation of caveolin-1 by mulberry leaf extract and its major components, chlorogenic acid derivatives, attenuates alcoholic steatohepatitis via inhibition of oxidative stress. Food Funct. 2017, 8, 397–405. [Google Scholar] [CrossRef]
- Peng, C.-H.; Lin, H.-T.; Chung, D.-J.; Huang, C.-N.; Wang, C.-J. Mulberry Leaf Extracts prevent obesity-induced NAFLD with regulating adipocytokines, inflammation and oxidative stress. J. Food Drug Anal. 2018, 26, 778–787. [Google Scholar] [CrossRef]
- Ann, J.-Y.; Eo, H.; Lim, Y. Mulberry leaves (Morus alba L.) ameliorate obesity-induced hepatic lipogenesis, fibrosis, and oxidative stress in high-fat diet-fed mice. Genes Nutr. 2015, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.R.; Kim, J.E.; Park, J.W.; Kang, M.J.; Choi, H.J.; Bae, S.J.; Choi, Y.W.; Kim, K.M.; Hong, J.T.; Hwang, D.Y. Fermented mulberry (Morus alba) leaves suppress high fat diet-induced hepatic steatosis through amelioration of the inflammatory response and autophagy pathway. BMC Complementary Med. Ther. 2020, 20, 283. [Google Scholar] [CrossRef]
- Bajko, E.; Kalinowska, M.; Borowski, P.; Siergiejczyk, L.; Lewandowski, W. 5-O-Caffeoylquinic acid: A spectroscopic study and biological screening for antimicrobial activity. LWT-Food Sci. Technol. 2016, 65, 471–479. [Google Scholar] [CrossRef]
- Hao, S.; Xiao, Y.; Lin, Y.; Mo, Z.; Chen, Y.; Peng, X.; Xiang, C.; Li, Y.; Li, W. Chlorogenic acid-enriched extract from Eucommia ulmoides leaves inhibits hepatic lipid accumulation through regulation of cholesterol metabolism in HepG2 cells. Pharm. Biol. 2016, 54, 251–259. [Google Scholar] [CrossRef]
- Tsai, K.L.; Hung, C.H.; Chan, S.H.; Hsieh, P.L.; Ou, H.C.; Cheng, Y.H.; Chu, P.M. Chlorogenic acid protects against oxLDL-induced oxidative damage and mitochondrial dysfunction by modulating SIRT1 in endothelial cells. Mol. Nutr. Food Res. 2018, 62, 1700928. [Google Scholar] [CrossRef]
- Randy, A.; Kim, M.; Nho, C.W. Ligularia fischeri and its constituent 3, 4-dicaffeoylquinic acid improve obesity-induced nonalcoholic fatty liver disease by regulating lipid metabolism and activating AMPK. J. Funct. Foods 2016, 27, 1–16. [Google Scholar] [CrossRef]
- Huang, K.; Liang, X.c.; Zhong, Y.l.; He, W.y.; Wang, Z. 5-Caffeoylquinic acid decreases diet-induced obesity in rats by modulating PPARα and LXRα transcription. J. Sci. Food Agric. 2015, 95, 1903–1910. [Google Scholar] [CrossRef]
- Kim, D.; Touros, A.; Kim, W.R. Nonalcoholic fatty liver disease and metabolic syndrome. Clin. Liver Dis. 2018, 22, 133–140. [Google Scholar] [CrossRef]
- Shi, L.; Karrar, E.; Wang, X. Sesamol ameliorates hepatic lipid accumulation and oxidative stress in steatosis HepG2 cells via the PPAR signaling pathway. J. Food Biochem. 2021, 45, e13976. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, J.; Wang, W.; Zhang, L.; Xu, J.; Wang, K.; Li, D. Resveratrol supplement inhibited the NF-κB inflammation pathway through activating AMPKα-SIRT1 pathway in mice with fatty liver. Mol. Cell. Biochem. 2016, 422, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular actions of PPAR α in lipid metabolism and inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Sun, M.; Cao, Y.; Ma, L.; Shen, Y.; Velikanova, A.A.; Li, X.; Sun, C.; Zhao, Y. miR-34a regulates lipid metabolism by targeting SIRT1 in non-alcoholic fatty liver disease with iron overload. Arch. Biochem. Biophys. 2020, 695, 108642. [Google Scholar] [CrossRef]
- Parlati, L.; Régnier, M.; Guillou, H.; Postic, C. New targets for NAFLD. JHEP Rep. 2021, 3, 100346. [Google Scholar] [CrossRef]
- Leoni, S.; Tovoli, F.; Napoli, L.; Serio, I.; Ferri, S.; Bolondi, L. Current guidelines for the management of non-alcoholic fatty liver disease: A systematic review with comparative analysis. World J. Gastroenterol. 2018, 24, 3361. [Google Scholar] [CrossRef]
- Yadav, A.; Shah, N.; Tiwari, P.K.; Javed, K.; Cheng, Q.; Aidhen, I.S.; Bröer, S. Novel chemical scaffolds to inhibit the neutral amino acid transporter B0AT1 (SLC6A19), a potential target to treat metabolic diseases. Front. Pharmacol. 2020, 11, 140. [Google Scholar] [CrossRef]
- Rafiei, H.; Omidian, K.; Bandy, B. Dietary polyphenols protect against oleic acid-induced steatosis in an in vitro model of NAFLD by modulating lipid metabolism and improving mitochondrial function. Nutrients 2019, 11, 541. [Google Scholar] [CrossRef] [Green Version]
- Sui, G.-G.; Xiao, H.-B.; Lu, X.-Y.; Sun, Z.-L. Naringin activates AMPK resulting in altered expression of SREBPs, PCSK9, and LDLR to reduce body weight in obese C57BL/6J mice. J. Agric. Food Chem. 2018, 66, 8983–8990. [Google Scholar] [CrossRef]
- Blade, C.; Baselga-Escudero, L.; Arola-Arnal, A. microRNAs as new targets of dietary polyphenols. Curr. Pharm. Biotechnol. 2014, 15, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Tajner-Czopek, A.; Gertchen, M.; Rytel, E.; Kita, A.; Kucharska, A.Z.; Sokół-Łętowska, A. Study of Antioxidant Activity of Some Medicinal Plants Having High Content of Caffeic Acid Derivatives. Antioxidants 2020, 9, 412. [Google Scholar] [CrossRef]
- Wang, S.; Sarriá, B.; Mateos, R.; Goya, L.; Bravo-Clemente, L. TNF-α-induced oxidative stress and endothelial dysfunction in EA. hy926 cells is prevented by mate and green coffee extracts, 5-caffeoylquinic acid and its microbial metabolite, dihydrocaffeic acid. Int. J. Food Sci. Nutr. 2019, 70, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Santana-Gálvez, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Chlorogenic acid: Recent advances on its dual role as a food additive and a nutraceutical against metabolic syndrome. Molecules 2017, 22, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Padhye, A.; Sharma, A.; Song, G.; Miao, J.; Mo, Y.-Y.; Wang, L.; Kemper, J.K. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 2010, 285, 12604–12611. [Google Scholar] [CrossRef] [Green Version]
- Flowers, E.; Froelicher, E.S.; Aouizerat, B.E. MicroRNA regulation of lipid metabolism. Metabolism 2013, 62, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Salvoza, N.C.; Klinzing, D.C.; Gopez-Cervantes, J.; Baclig, M.O. Association of circulating serum miR-34a and miR-122 with dyslipidemia among patients with non-alcoholic fatty liver disease. PLoS ONE 2016, 11, e0153497. [Google Scholar] [CrossRef] [PubMed]
- Majeed, Y.; Halabi, N.; Madani, A.Y.; Engelke, R.; Bhagwat, A.M.; Abdesselem, H.; Agha, M.V.; Vakayil, M.; Courjaret, R.; Goswami, N. SIRT1 promotes lipid metabolism and mitochondrial biogenesis in adipocytes and coordinates adipogenesis by targeting key enzymatic pathways. Sci. Rep. 2021, 11, 8177. [Google Scholar] [CrossRef]
- Li, X.; Lian, F.; Liu, C.; Hu, K.-Q.; Wang, X.-D. Isocaloric pair-fed high-carbohydrate diet induced more hepatic steatosis and inflammation than high-fat diet mediated by miR-34a/SIRT1 axis in mice. Sci. Rep. 2015, 5, 16774. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xu, S.; Maitland-Toolan, K.A.; Sato, K.; Jiang, B.; Ido, Y.; Lan, F.; Walsh, K.; Wierzbicki, M.; Verbeuren, T.J. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 2008, 283, 20015–20026. [Google Scholar] [CrossRef] [Green Version]
- Meng, S.; Cao, J.; Feng, Q.; Peng, J.; Hu, Y. Roles of chlorogenic acid on regulating glucose and lipids metabolism: A review. Evid.-Based Complementary Altern. Med. 2013, 2013, 801457. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, M.; Lü, J. Determination of rutin, quercetin, and chlorogenic acid in mulberry leaves by capillary zone electrophoresis. Anal. Lett. 2004, 37, 3287–3297. [Google Scholar] [CrossRef]
- Zhai, K.-F.; Duan, H.; Shi, S.-X.; Liu, L.-L.; Cao, W.-G.; Gao, G.-Z.; Shan, L.-L. Synchronised determination of chlorogenic acid and five flavonoids in mulberry leaves using HPLC with photodiode array detection. Qual. Assur. Saf. Crop. Foods 2018, 10, 175–182. [Google Scholar] [CrossRef]
- Liang, N.; Dupuis, J.H.; Yada, R.Y.; Kitts, D.D. Chlorogenic acid isomers directly interact with Keap 1-Nrf2 signaling in Caco-2 cells. Mol. Cell. Biochem. 2019, 457, 105–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, M.; Rheinheimer, S.; Leidinger, P.; Backes, C.; Menegatti, J.; Fehlmann, T.; Grässer, F.; Keller, A.; Meese, E. Identification of miR-34a-target interactions by a combined network based and experimental approach. Oncotarget 2016, 7, 34288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, F.; Yang, J.; Li, Q.; Zhou, H. Down-regulation of microRNA-34a-5p promotes trophoblast cell migration and invasion via targetting Smad4. Biosci. Rep. 2019, 39, BSR20181631. [Google Scholar] [CrossRef] [PubMed] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.-H.; Hung, T.-W.; Wang, C.-C.; Wu, S.-W.; Yang, T.-W.; Yang, C.-Y.; Tseng, T.-H.; Wang, C.-J. Neochlorogenic Acid Attenuates Hepatic Lipid Accumulation and Inflammation via Regulating miR-34a In Vitro. Int. J. Mol. Sci. 2021, 22, 13163. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222313163
Yu M-H, Hung T-W, Wang C-C, Wu S-W, Yang T-W, Yang C-Y, Tseng T-H, Wang C-J. Neochlorogenic Acid Attenuates Hepatic Lipid Accumulation and Inflammation via Regulating miR-34a In Vitro. International Journal of Molecular Sciences. 2021; 22(23):13163. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222313163
Chicago/Turabian StyleYu, Meng-Hsun, Tung-Wei Hung, Chi-Chih Wang, Sheng-Wen Wu, Tzu-Wei Yang, Ching-Yu Yang, Tsui-Hwa Tseng, and Chau-Jong Wang. 2021. "Neochlorogenic Acid Attenuates Hepatic Lipid Accumulation and Inflammation via Regulating miR-34a In Vitro" International Journal of Molecular Sciences 22, no. 23: 13163. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222313163