Differential Regulation of Bilastine Affinity for Human Histamine H1 Receptors by Lys 179 and Lys 191 via Its Binding Enthalpy and Entropy
Abstract
:1. Introduction
2. Results and Discussion
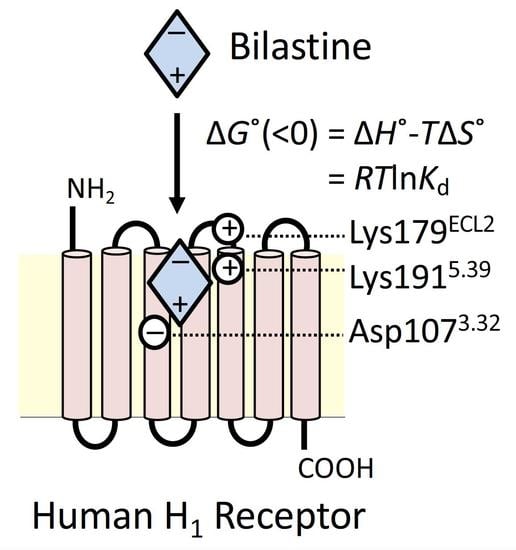
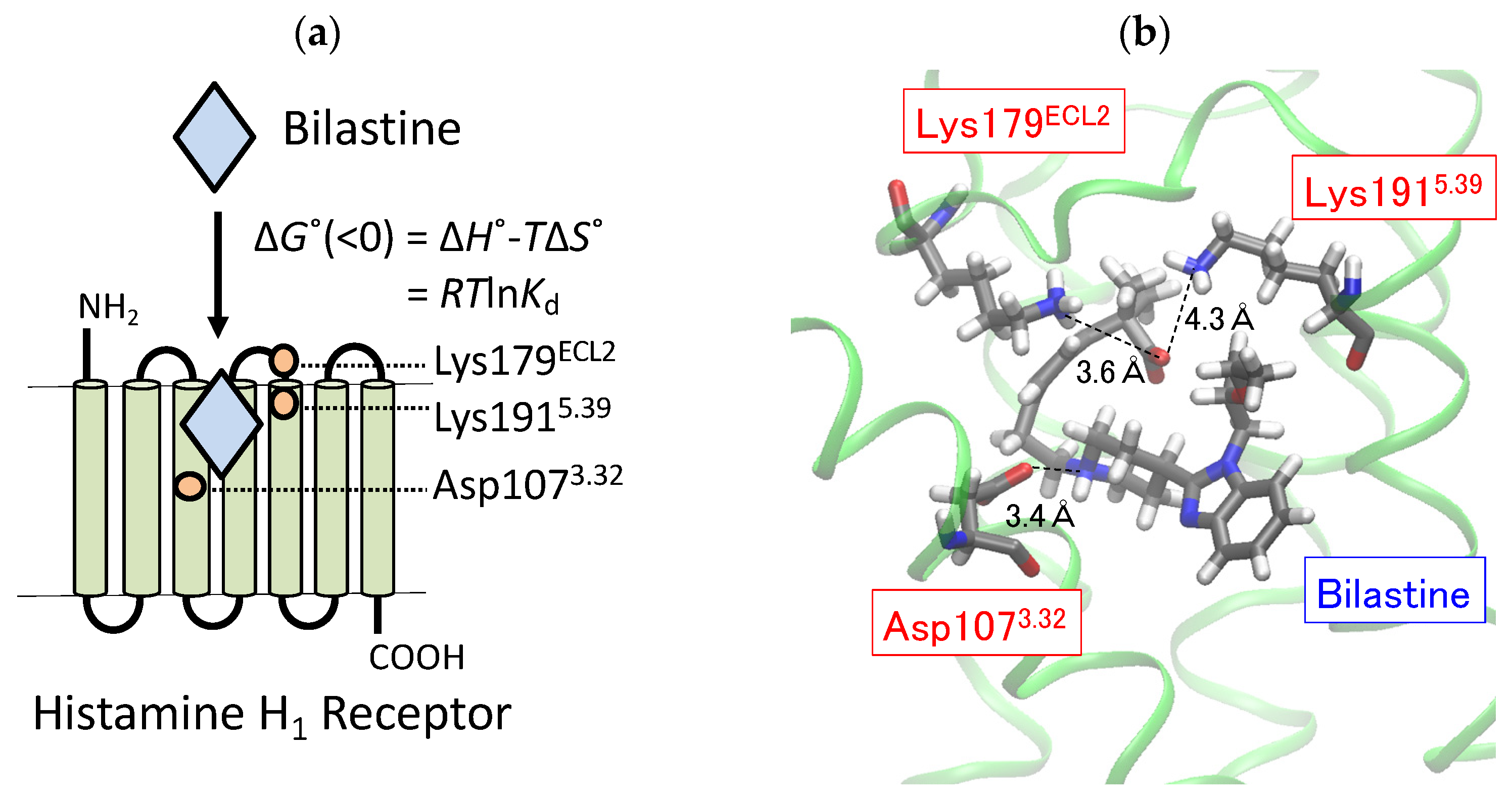
2.1. Docking Simulation on the Binding of Bilastine to H1 Receptor
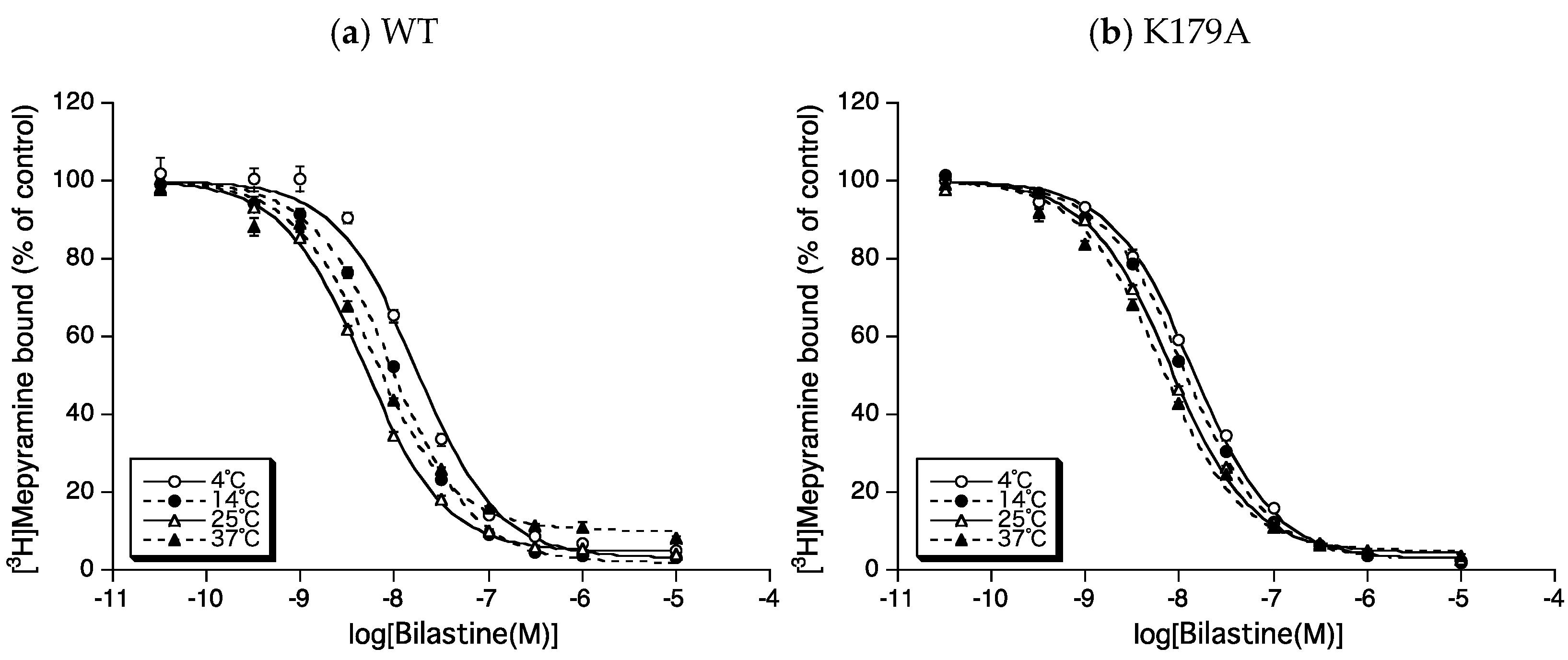
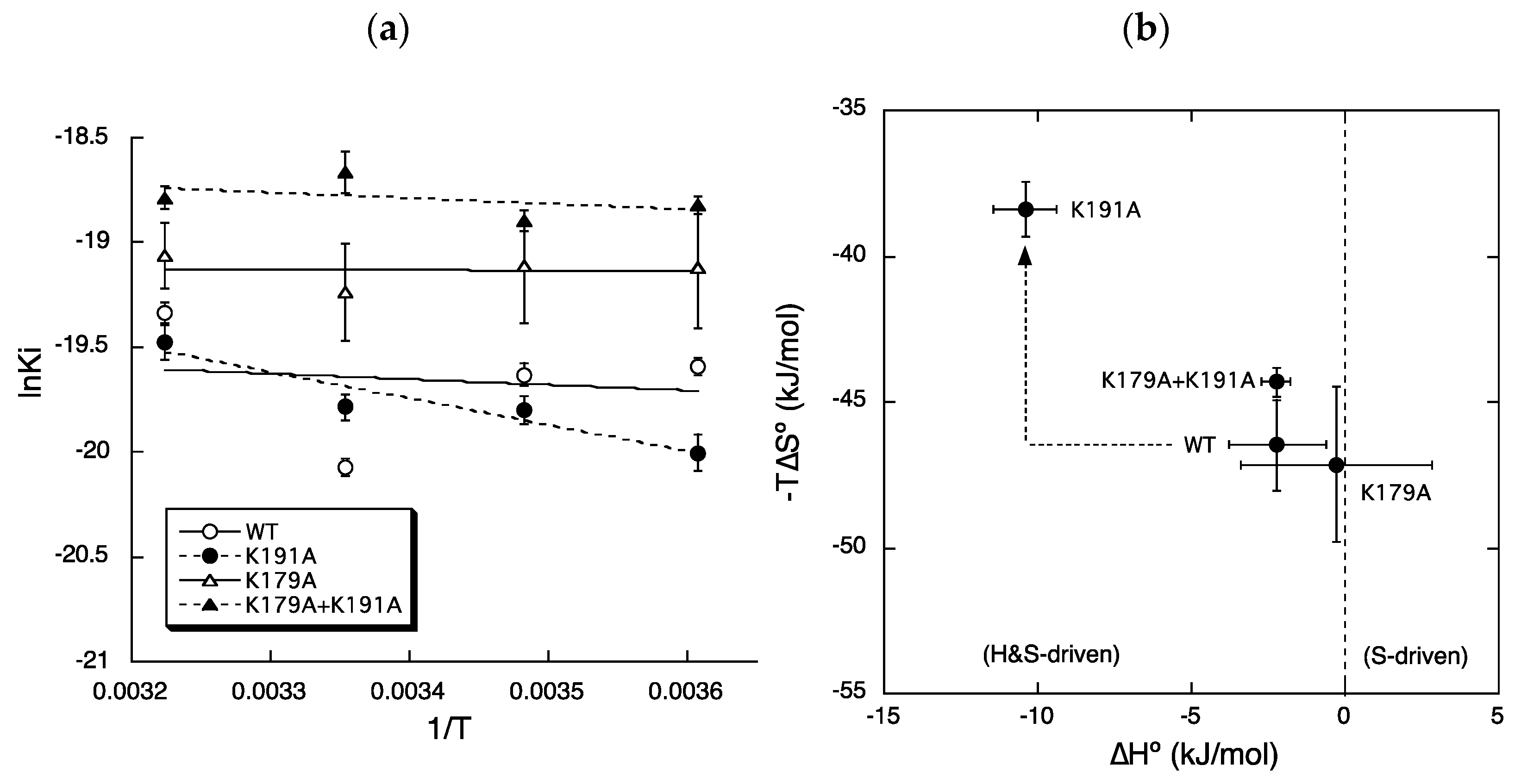
2.2. Roles of Lys179ECL2 and Lys1915.39 in the Binding Affinity for Bilastine
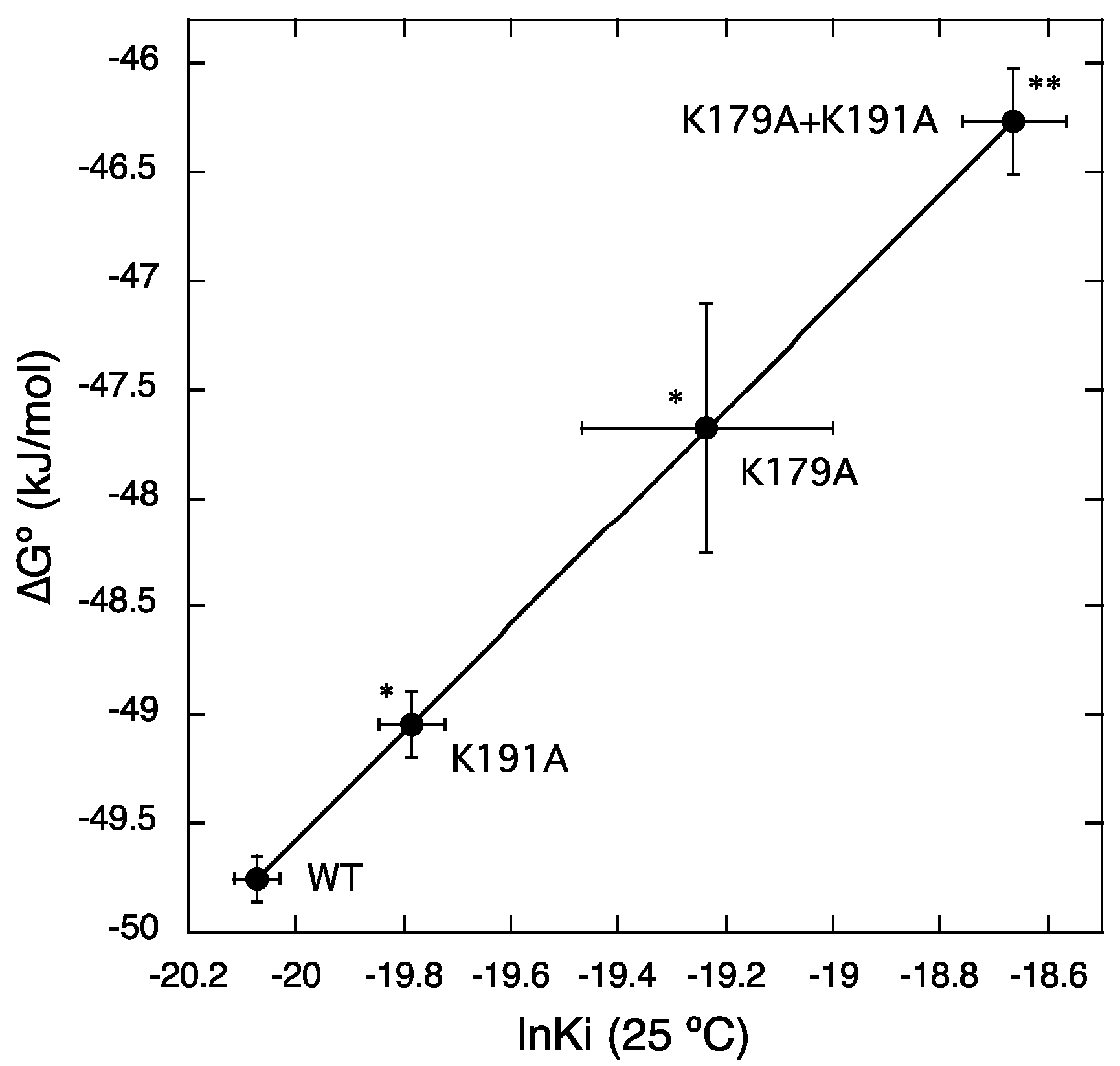
2.3. Roles of Lys179ECL2 and Lys1915.39 in Thermodynamic Binding Forces of Bilastine
3. Materials and Methods
3.1. Materials
3.2. Docking Simulation on the Binding of Bilastine to Human H1 Receptor
3.3. Measurement of [3H]Mepyramine Binding to Membrane Preparations
3.4. Data Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CHO | Chinese hamster ovary |
| K179A | Lys179 mutant of H1 receptor to alanine |
| K191A | Lys191 mutant of H1 receptor to alanine |
| K179A + K191A | Both Lys179 and Lys191 mutant of H1 receptor to alanine |
| WT | Wild-type H1 receptor |
References
- Hill, S.J. Distribution, properties, and functional characteristics of three classes of histamine receptor. Pharmacol. Rev. 1990, 42, 45–83. [Google Scholar] [PubMed]
- Hill, S.J.; Ganellin, C.R.; Timmerman, H.; Schwartz, J.C.; Shankley, N.P.; Young, J.M.; Schunack, W.; Levi, R.; Haas, H.L. International Union of Pharmacology. XIII. Classification of histamine receptors. Pharmacol. Rev. 1997, 49, 253–278. [Google Scholar] [PubMed]
- Smit, M.J.; Hoffmann, M.; Timmerman, H.; Leurs, R. Molecular properties and signalling pathways of the histamine H1 receptor. Clin. Exp. Allergy 1999, 29 (Suppl. 3), 19–28. [Google Scholar] [CrossRef] [PubMed]
- Bakker, R.A.; Timmerman, H.; Leurs, R. Histamine receptors: Specific ligands, receptor biochemistry, and signal transduction. Clin. Allergy Immunol. 2002, 17, 27–64. [Google Scholar]
- Yanai, K.; Tashiro, M. The physiological and pathophysiological roles of neuronal histamine: An insight from human positron emission tomography studies. Pharmacol. Ther. 2007, 113, 1–15. [Google Scholar] [CrossRef]
- Haas, H.L.; Sergeeva, O.A.; Selbach, O. Histamine in the nervous system. Physiol. Rev. 2008, 88, 1183–1241. [Google Scholar] [CrossRef]
- Sadek, B.; Stark, H. Cherry-picked ligands at histamine receptor subtypes. Neuropharmacology 2016, 106, 56–73. [Google Scholar] [CrossRef]
- Tiligada, E.; Ennis, M. Histamine pharmacology: From Sir Henry Dale to the 21st century. Br. J. Pharmacol. 2020, 177, 469–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holgate, S.T.; Canonica, G.W.; Simons, F.E.; Taglialatela, M.; Tharp, M.; Timmerman, H.; Yanai, K. Consensus group on new-generation antihistamines (CONGA): Present status and recommendations. Clin. Exp. Allergy 2003, 33, 1305–1324. [Google Scholar] [CrossRef] [Green Version]
- Kalpaklioglu, F.; Baccioglu, A. Efficacy and safety of H1-antihistamines: An update. Antiinflamm. Antiallergy Agents Med. Chem. 2012, 11, 230–237. [Google Scholar] [CrossRef]
- Yanai, K.; Yoshikawa, T.; Yanai, A.; Nakamura, T.; Iida, T.; Leurs, R.; Tashiro, M. The clinical pharmacology of non-sedating antihistamines. Pharmacol. Ther. 2017, 178, 148–156. [Google Scholar] [CrossRef]
- Church, M.K. Allergy, histamine and antihistamines. Handb. Exp. Pharmacol. 2017, 241, 321–331. [Google Scholar] [PubMed]
- Kawauchi, H.; Yanai, K.; Wang, D.Y.; Itahashi, K.; Okubo, K. Antihistamines for Allergic Rhinitis Treatment from the Viewpoint of Nonsedative Properties. Int. J. Mol. Sci. 2019, 20, 213. [Google Scholar] [CrossRef] [Green Version]
- Farré, M.; Pérez-Mañá, C.; Papaseit, E.; Menoyo, E.; Pérez, M.; Martin, S.; Bullich, S.; Rojas, S.; Herance, J.R.; Corcóstegui, R.; et al. Preclinical pharmacology of bilastine, a new selective histamine H1 receptor antagonist: Receptor selectivity and in vitro antihistaminic activity. Drugs R. D. 2005, 6, 371–384. [Google Scholar]
- Church, M.K. Safety and efficacy of bilastine: A new H1-antihistamine for the treatment of allergic rhinoconjunctivitis and urticaria. Expert Opin. Drug Saf. 2011, 10, 779–793. [Google Scholar] [CrossRef]
- Farré, M.; Pérez-Mañá, C.; Papaseit, E.; Menoyo, E.; Pérez, M.; Martin, S.; Bullich, S.; Rojas, S.; Herance, J.R.; Trampal, C.; et al. Bilastine vs. hydroxyzine: Occupation of brain histamine H1-receptors evaluated by positron emission tomography in healthy volunteers. Br. J. Clin. Pharmacol. 2014, 78, 970–980. [Google Scholar]
- Wang, X.Y.; Lim-Jurado, M.; Prepageran, N.; Tantilipikorn, P.; Wang de, Y. Treatment of allergic rhinitis and urticaria: A review of the newest antihistamine drug bilastine. Ther. Clin. Risk Manag. 2016, 12, 585–597. [Google Scholar] [CrossRef] [Green Version]
- Bosma, R.; van den Bor, J.; Vischer, H.F.; Labeaga, L.; Leurs, R. The long duration of action of the second generation antihistamine bilastine coincides with its long residence time at the histamine H1 receptor. Eur. J. Pharmacol. 2018, 838, 107–111. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Wakugawa, T.; Sadakata, H.; Kamimura, S.; Takemoto, M.; Nakagawa, T.; Yabumoto, M.; Kitamura, Y.; Takeda, N.; Fukui, H. Elucidation of Inverse Agonist Activity of Bilastine. Pharmaceutics 2020, 12, 525. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Shimamura, T.; Shiroishi, M.; Weyand, S.; Tsujimoto, H.; Winter, G.; Katritch, V.; Abagyan, R.; Cherezov, V.; Liu, W.; Han, G.W.; et al. Structure of the human histamine H1 receptor complex with doxepin. Nature 2011, 475, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Hitzemann, R. Thermodynamics aspects of drug–receptor interactions. Trends Pharm. Sci. 1988, 9, 408–411. [Google Scholar] [CrossRef]
- Borea, P.A.; Dalpiaz, A.; Varani, K.; Gilli, P.; Gilli, G. Can thermodynamic measurements of receptor binding yield information on drug affinity and efficacy? Biochem. Pharmacol. 2000, 60, 1549–1556. [Google Scholar] [CrossRef]
- Wittmann, H.J.; Seifert, R.; Strasser, A. Contribution of binding enthalpy and entropy to affinity of antagonist and agonist binding at human and guinea pig histamine H1-receptor. Mol. Pharmacol. 2009, 76, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Hishinuma, S.; Sugawara, K.; Uesawa, Y.; Fukui, H.; Shoji, M. Differential thermodynamic driving force of first- and second-generation antihistamines to determine their binding affinity for human H1 receptors. Biochem. Pharmacol. 2014, 91, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Hishinuma, S.; Tamura, Y.; Kobayashi, C.; Akatsu, C.; Shoji, M. Differential regulation of thermodynamic binding forces of levocetirizine and (S)-cetirizine by Lys191 in human histamine H₁ receptors. Int. J. Mol. Sci. 2018, 19, 4067. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, C.; Tanaka, A.; Yasuda, T.; Hishinuma, S. Roles of Lys191 and Lys179 in regulating thermodynamic binding forces of ligands to determine their binding affinity for human histamine H1 receptors. Biochem. Pharmacol. 2020, 180, 114185. [Google Scholar] [CrossRef] [PubMed]
- Chandler, D. Interfaces and the driving force of hydrophobic assembly. Nature 2005, 437, 640–647. [Google Scholar] [CrossRef]
- Gilman-Politi, R.; Harries, D. Unraveling the Molecular Mechanism of Enthalpy Driven Peptide Folding by Polyol Osmolytes. J. Chem. Theory Comput. 2011, 7, 3816–3828. [Google Scholar] [CrossRef] [PubMed]
- Schauperl, M.; Podewitz, M.; Waldner, B.J.; Liedl, K.R. Enthalpic and Entropic Contributions to Hydrophobicity. J. Chem. Theory Comput. 2016, 12, 4600–4610. [Google Scholar] [CrossRef]
- Fukunishi, Y.; Mikami, Y.; Nakamura,, H. Similarities among receptor pockets and among compounds: Analysis and application to in silico ligand screening. J. Mol. Graph. Model. 2005, 24, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Vigers, G.P.; Rizzi, J.P. Multiple active site corrections for docking and virtual screening. J. Med. Chem. 2004, 47, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Lamdan, Y.; Schwartz, J.; Wolfson, H. Affine invariant model-based object recognition. IEEE Trans. Robot. Automat. 1990, 6, 578–589. [Google Scholar] [CrossRef]
- Chen, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akimoto, H.; Sugihara, M.; Hishinuma, S. Differential Regulation of Bilastine Affinity for Human Histamine H1 Receptors by Lys 179 and Lys 191 via Its Binding Enthalpy and Entropy. Int. J. Mol. Sci. 2021, 22, 1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041655
Akimoto H, Sugihara M, Hishinuma S. Differential Regulation of Bilastine Affinity for Human Histamine H1 Receptors by Lys 179 and Lys 191 via Its Binding Enthalpy and Entropy. International Journal of Molecular Sciences. 2021; 22(4):1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041655
Chicago/Turabian StyleAkimoto, Hayato, Minoru Sugihara, and Shigeru Hishinuma. 2021. "Differential Regulation of Bilastine Affinity for Human Histamine H1 Receptors by Lys 179 and Lys 191 via Its Binding Enthalpy and Entropy" International Journal of Molecular Sciences 22, no. 4: 1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041655