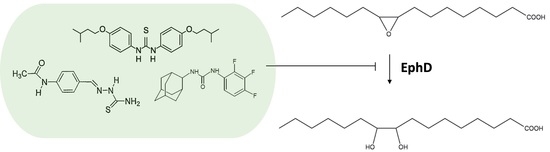
Mycobacterial Epoxide Hydrolase EphD Is Inhibited by Urea and Thiourea Derivatives
Abstract
:1. Introduction
2. Results
2.1. Monitoring the Activity of Mycobacterial EHs in the Presence of ISO
2.2. EphD and EphF Interact Differently with ISO
2.3. EphD Is Inhibited by Urea and Thiourea Derivatives inside the Cell
3. Discussion
4. Materials and Methods
4.1. Cloning, Bacterial Strains, Growth Conditions and Recombinant Protein Production
4.2. Purification of EthA
4.3. Epoxide Hydrolase Assays
4.4. Analysis of Mycolic Acids
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Huszár, S.; Chibale, K.; Singh, V. The quest for the holy grail: New antitubercular chemical entities, targets and strategies. Drug Discov. Today 2020, 25, 772–780. [Google Scholar] [CrossRef]
- Kroesen, V.M.; Madacki, J.; Frigui, W.; Sayes, F.; Brosch, R. Mycobacterial virulence: Impact on immunogenicity and vaccine research. F1000Research 2019, 8, 2025. [Google Scholar] [CrossRef]
- McShane, H. Insights and challenges in tuberculosis vaccine development. Lancet Respir. Med. 2019, 7, 810–819. [Google Scholar] [CrossRef]
- WHO. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment; World Health Organization: Geneva, Switzerland, 2019; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Tousek, J. On the Clinical effectiveness of isoxyl. Antibiot. Chemother. 1970, 16, 149–155. [Google Scholar] [PubMed]
- Phetsuksiri, B.; Baulard, A.R.; Cooper, A.M.; Minnikin, D.E.; Douglas, J.D.; Besra, G.S.; Brennan, P.J. Antimycobacterial activities of isoxyl and new derivatives through the inhibition of mycolic acid synthesis. Antimicrob. Agents Chemother. 1999, 43, 1042–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phetsuksiri, B.; Jackson, M.; Scherman, H.; McNeil, M.; Besra, G.S.; Baulard, A.R.; Slayden, R.A.; DeBarber, A.E.; Barry, C.E.; Baird, M.S.; et al. Unique mechanism of action of the thiourea drug isoxyl on mycobacterium tuberculosis. J. Biol. Chem. 2003, 278, 53123–53130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portevin, D.; de Sousa-D’Auria, C.; Houssin, C.; Grimaldi, C.; Chami, M.; Daffé, M.; Guilhot, C. A Polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. USA 2004, 101, 314–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilchèze, C.; Morbidoni, H.R.; Weisbrod, T.R.; Iwamoto, H.; Kuo, M.; Sacchettini, J.C.; Jacobs, W.R. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of mycobacterium smegmatis. J. Bacteriol. 2000, 182, 4059–4067. [Google Scholar] [CrossRef] [Green Version]
- Barry, C.E., III; Lee, R.E.; Mdluli, K.; Sampson, A.E.; Schroeder, B.G.; Slayden, R.A.; Yuan, Y. Mycolic acids: Structure, biosynthesis and physiological functions. Prog. Lipid Res. 1998, 37, 143–179. [Google Scholar] [CrossRef] [Green Version]
- Grzegorzewicz, A.E.; Korduláková, J.; Jones, V.; Born, S.E.M.; Belardinelli, J.M.; Vaquié, A.; Gundi, V.A.K.B.; Madacki, J.; Slama, N.; Laval, F.; et al. A common mechanism of inhibition of the mycobacterium tuberculosis mycolic acid biosynthetic pathway by isoxyl and thiacetazone. J. Biol. Chem. 2012, 287, 38434–38441. [Google Scholar] [CrossRef] [Green Version]
- Dover, L.G.; Alahari, A.; Gratraud, P.; Gomes, J.M.; Bhowruth, V.; Reynolds, R.C.; Besra, G.S.; Kremer, L. EthA, a common activator of thiocarbamide-containing drugs acting on different mycobacterial targets. Antimicrob. Agents Chemother. 2007, 51, 1055–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzegorzewicz, A.E.; Eynard, N.; Quémard, A.; North, E.J.; Margolis, A.; Lindenberger, J.J.; Jones, V.; Korduláková, J.; Brennan, P.J.; Lee, R.E.; et al. Covalent modification of the mycobacterium tuberculosis FAS-II dehydratase by isoxyl and thiacetazone. ACS Infect. Dis. 2015, 1, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Korduláková, J.; Janin, Y.L.; Liav, A.; Barilone, N.; Vultos, T.D.; Rauzier, J.; Brennan, P.J.; Gicquel, B.; Jackson, M. Isoxyl activation is required for bacteriostatic activity against mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 3824–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daffé, M.; Marrakchi, H. Unraveling the structure of the mycobacterial envelope. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Hoffmann, C.; Leis, A.; Niederweis, M.; Plitzko, J.M.; Engelhardt, H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 2008, 105, 3963–3967. [Google Scholar] [CrossRef] [Green Version]
- Zuber, B.; Chami, M.; Houssin, C.; Dubochet, J.; Griffiths, G.; Daffé, M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008, 190, 5672–5680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; de Lisle, G.; Jacobs, W.R. InhA, a gene encoding a target for isoniazid and ethionamide in mycobacterium tuberculosis. Science 1994, 263, 227–230. [Google Scholar] [CrossRef] [Green Version]
- Takayama, K.; Wang, L.; David, H.L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1972, 2, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Belanger, A.E.; Besra, G.S.; Ford, M.E.; Mikusová, K.; Belisle, J.T.; Brennan, P.J.; Inamine, J.M. The EmbAB genes of mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 1996, 93, 11919–11924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzegorzewicz, A.E.; Pham, H.; Gundi, V.A.K.B.; Scherman, M.S.; North, E.J.; Hess, T.; Jones, V.; Gruppo, V.; Born, S.E.M.; Korduláková, J.; et al. Inhibition of mycolic acid transport across the mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 2012, 8, 334–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belisle, J.T.; Vissa, V.D.; Sievert, T.; Takayama, K.; Brennan, P.J.; Besra, G.S. Role of the major antigen of mycobacterium tuberculosis in cell wall biogenesis. Science 1997, 276, 1420–1422. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the biology of mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Tekaia, F.; Gordon, S.V.; Garnier, T.; Brosch, R.; Barrell, B.G.; Cole, S.T. Analysis of the proteome of mycobacterium tuberculosis in silico. Tuber. Lung Dis. 1999, 79, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Johansson, P.; Unge, T.; Cronin, A.; Arand, M.; Bergfors, T.; Jones, T.A.; Mowbray, S.L. Structure of an Atypical epoxide hydrolase from mycobacterium tuberculosis gives insights into its function. J. Mol. Biol. 2005, 351, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Madacki, J.; Laval, F.; Grzegorzewicz, A.; Lemassu, A.; Záhorszká, M.; Arand, M.; McNeil, M.; Daffé, M.; Jackson, M.; Lanéelle, M.-A.; et al. Impact of the epoxide hydrolase EphD on the metabolism of mycolic acids in mycobacteria. J. Biol. Chem. 2018, 293, 5172–5184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswal, B.K.; Garen, G.; Cherney, M.M.; Garen, C.; James, M.N.G. Cloning, expression, purification, crystallization and preliminary X-ray studies of epoxide hydrolases A and B from mycobacterium tuberculosis. Acta Cryst. F 2006, 62, 136–138. [Google Scholar] [CrossRef] [Green Version]
- Schulz, E.C.; Henderson, S.R.; Illarionov, B.; Crosskey, T.; Southall, S.M.; Krichel, B.; Uetrecht, C.; Fischer, M.; Wilmanns, M. The crystal structure of mycobacterial epoxide hydrolase A. Sci. Rep. 2020, 10, 16539. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.D.; Hammock, B.D. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat. Rev. Drug Discov. 2009, 8, 794–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morisseau, C.; Goodrow, M.H.; Dowdy, D.; Zheng, J.; Greene, J.F.; Sanborn, J.R.; Hammock, B.D. Potent urea and carbamate inhibitors of soluble epoxide hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 8849–8854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswal, B.K.; Morisseau, C.; Garen, G.; Cherney, M.M.; Garen, C.; Niu, C.; Hammock, B.D.; James, M.N.G. The molecular structure of epoxide hydrolase b from mycobacterium tuberculosis and its complex with a urea-based inhibitor. J. Mol. Biol. 2008, 381, 897–912. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.R.; North, E.J.; Hurdle, J.G.; Morisseau, C.; Scarborough, J.S.; Sun, D.; Korduláková, J.; Scherman, M.S.; Jones, V.; Grzegorzewicz, A.; et al. The structure–activity relationship of urea derivatives as anti-tuberculosis agents. Bioorganic Med. Chem. 2011, 19, 5585–5595. [Google Scholar] [CrossRef] [Green Version]
- Müller, F.; Arand, M.; Frank, H.; Seidel, A.; Hinz, W.; Winkler, L.; Hänel, K.; Blée, E.; Beetham, J.K.; Hammock, B.D.; et al. Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase, and their substrates. Eur. J. Biochem. 1997, 245, 490–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arand, M.; Cronin, A.; Adamska, M.; Oesch, F. Epoxide hydrolases: Structure, function, mechanism, and assay. In Phase II Conjugation Enzymes and Transport Systems; Sies, H., Packer, L., Eds.; Academic Press: Cambridge, MA, USA, 2005; Volume 400, pp. 569–588. ISBN 978-0-12-182805-9. [Google Scholar]
- Carrère-Kremer, S.; Blaise, M.; Singh, V.K.; Alibaud, L.; Tuaillon, E.; Halloum, I.; van de Weerd, R.; Guérardel, Y.; Drancourt, M.; Takiff, H.; et al. A New dehydratase conferring innate resistance to thiacetazone and intra-amoebal survival of mycobacterium smegmatis. Mol. Microbiol. 2015, 96, 1085–1102. [Google Scholar] [CrossRef] [PubMed]
- Triccas, J.A.; Parish, T.; Britton, W.J.; Gicquel, B. An inducible expression system permitting the efficient purification of a recombinant antigen from mycobacterium smegmatis. FEMS Microbiol. Lett. 1998, 167, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, B.; Kingma, J.; Arand, M.; Wubbolts, M.G.; Janssen, D.B. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl. Environ. Microbiol. 2006, 72, 2905–2917. [Google Scholar] [CrossRef] [Green Version]
- Gomez, G.A.; Morisseau, C.; Hammock, B.D.; Christianson, D.W. Human soluble epoxide hydrolase: Structural basis of inhibition by 4-(3-Cyclohexylureido)-carboxylic acids. Protein Sci. 2006, 15, 58–64. [Google Scholar] [CrossRef]
- Argiriadi, M.A.; Morisseau, C.; Hammock, B.D.; Christianson, D.W. Detoxification of environmental mutagens and carcinogens: Structure, mechanism, and evolution of liver epoxide hydrolase. Proc. Natl. Acad. Sci. USA 1999, 96, 10637–10642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argiriadi, M.A.; Morisseau, C.; Goodrow, M.H.; Dowdy, D.L.; Hammock, B.D.; Christianson, D.W. Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation. J. Biol. Chem. 2000, 275, 15265–15270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeJesus, M.A.; Gerrick, E.R.; Xu, W.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive essentiality analysis of the mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 2017, 8, e02133-16. [Google Scholar] [CrossRef] [Green Version]
- Rengarajan, J.; Bloom, B.R.; Rubin, E.J. Genome-wide requirements for mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 8327–8332. [Google Scholar] [CrossRef] [Green Version]
- Dao, D.N.; Sweeney, K.; Hsu, T.; Gurcha, S.S.; Nascimento, I.P.; Roshevsky, D.; Besra, G.S.; Chan, J.; Porcelli, S.A.; Jacobs, W.R. Mycolic acid modification by the MmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog. 2008, 4, e1000081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubnau, E.; Chan, J.; Raynaud, C.; Mohan, V.P.; Lanéelle, M.-A.; Yu, K.; Quémard, A.; Smith, I.; Daffé, M. Oxygenated mycolic acids are necessary for virulence of mycobacterium tuberculosis in mice. Mol. Microbiol. 2000, 36, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Bhowruth, V.; Brown, A.K.; Reynolds, R.C.; Coxon, G.D.; Mackay, S.P.; Minnikin, D.E.; Besra, G.S. Symmetrical and unsymmetrical analogues of isoxyl; active agents against mycobacterium tuberculosis. Bioorganic Med. Chem. Lett. 2006, 16, 4743–4747. [Google Scholar] [CrossRef] [PubMed]
- Coxon, G.D.; Craig, D.; Corrales, R.M.; Vialla, E.; Gannoun-Zaki, L.; Kremer, L. Synthesis, antitubercular activity and mechanism of resistance of highly effective thiacetazone analogues. PLoS ONE 2013, 8, e53162. [Google Scholar] [CrossRef] [Green Version]
- North, E.J.; Scherman, M.S.; Bruhn, D.F.; Scarborough, J.S.; Maddox, M.M.; Jones, V.; Grzegorzewicz, A.; Yang, L.; Hess, T.; Morisseau, C.; et al. Design, synthesis and anti-tuberculosis activity of 1-adamantyl-3-heteroaryl ureas with improved in vitro pharmacokinetic properties. Bioorganic Med. Chem. 2013, 21, 2587–2599. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madacki, J.; Kopál, M.; Jackson, M.; Korduláková, J. Mycobacterial Epoxide Hydrolase EphD Is Inhibited by Urea and Thiourea Derivatives. Int. J. Mol. Sci. 2021, 22, 2884. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22062884
Madacki J, Kopál M, Jackson M, Korduláková J. Mycobacterial Epoxide Hydrolase EphD Is Inhibited by Urea and Thiourea Derivatives. International Journal of Molecular Sciences. 2021; 22(6):2884. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22062884
Chicago/Turabian StyleMadacki, Jan, Martin Kopál, Mary Jackson, and Jana Korduláková. 2021. "Mycobacterial Epoxide Hydrolase EphD Is Inhibited by Urea and Thiourea Derivatives" International Journal of Molecular Sciences 22, no. 6: 2884. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22062884