Id2 Represses Aldosterone-Stimulated Cardiac T-Type Calcium Channels Expression
Abstract
:1. Introduction
2. Results
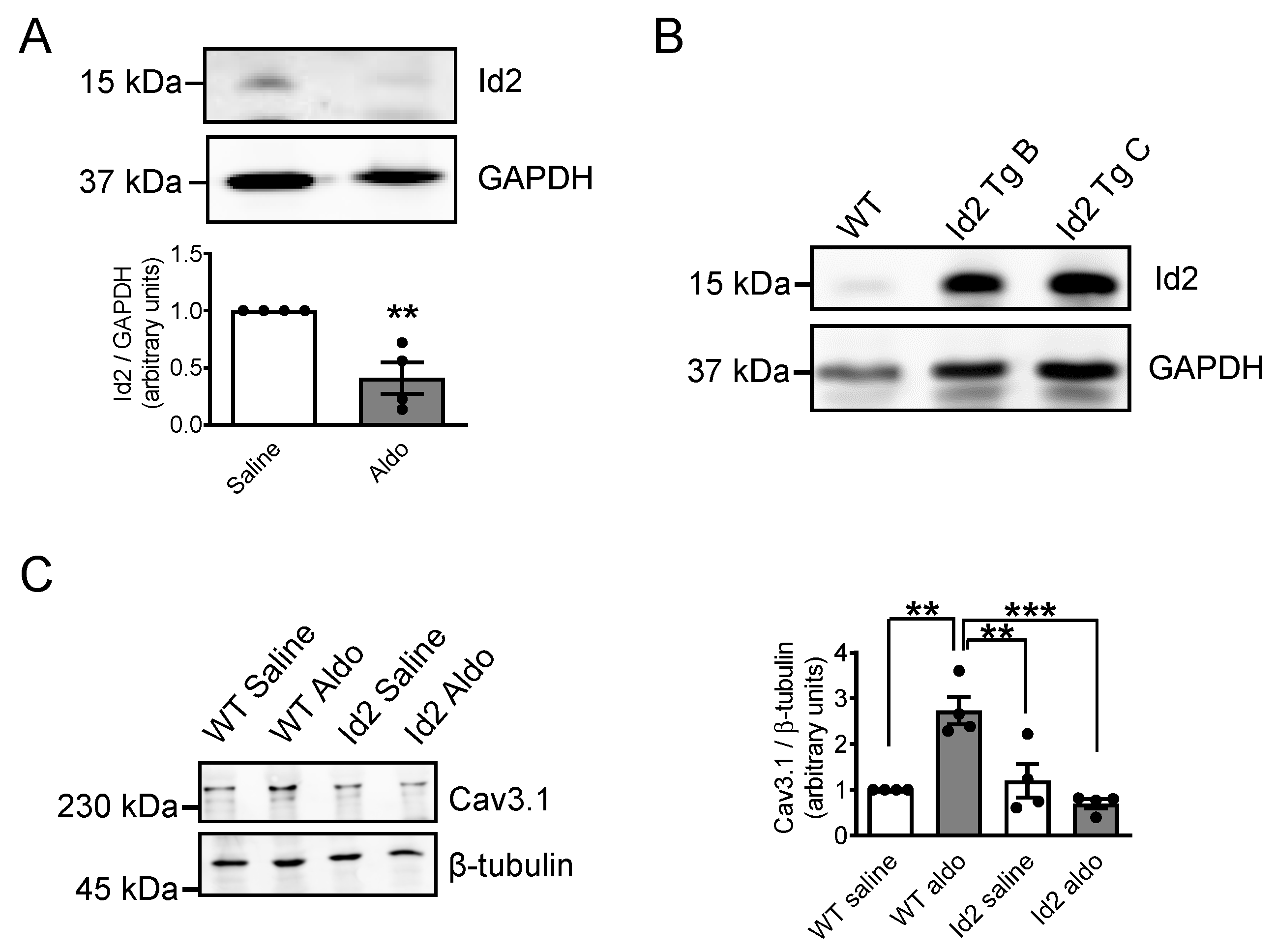
2.1. Aldosterone Represses Id2 Expression in Neonatal Rat Cardiomyocytes
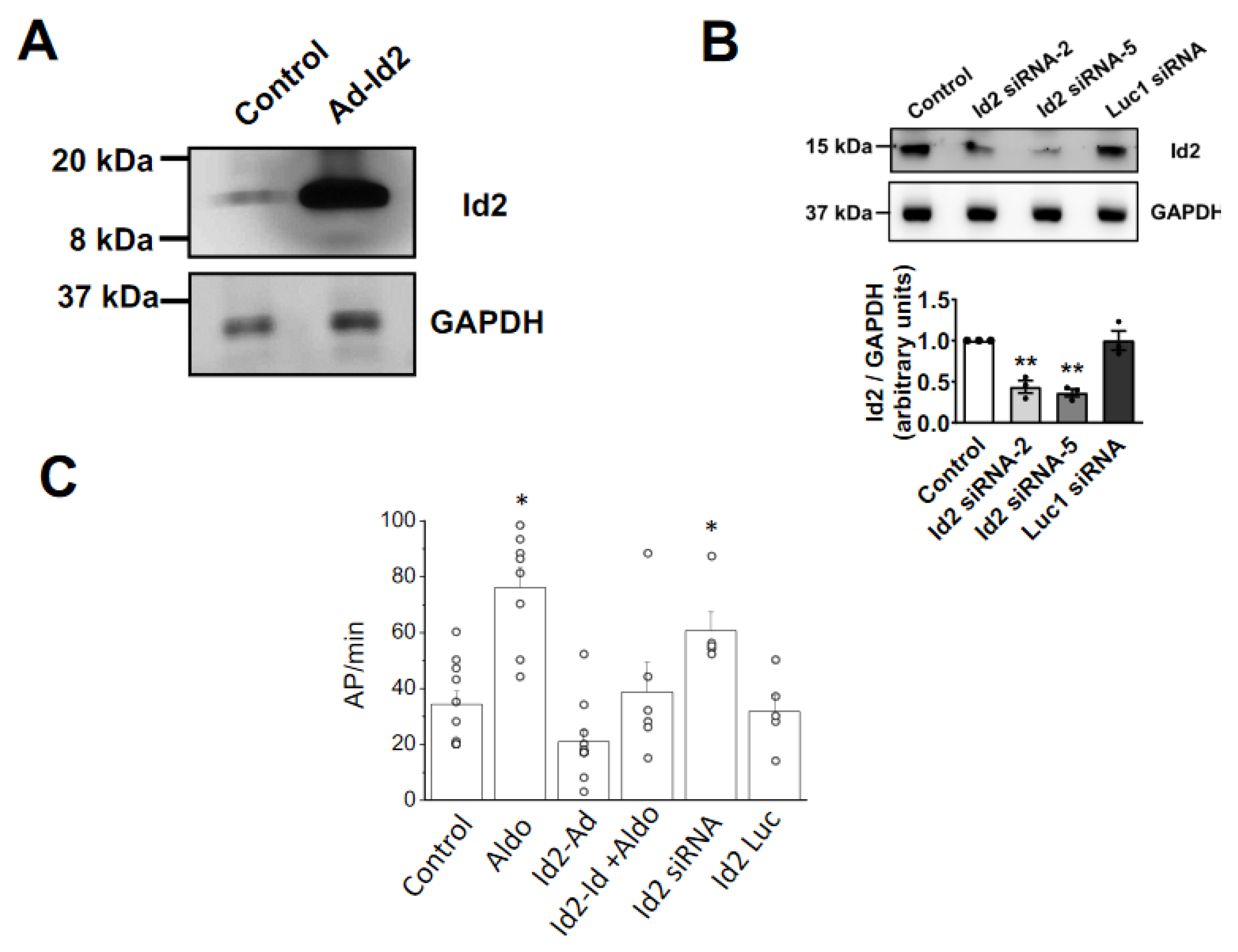
2.2. Overexpression of Id2 Prevents the Aldosterone-Stimulated Effects on Cardiomyocytes
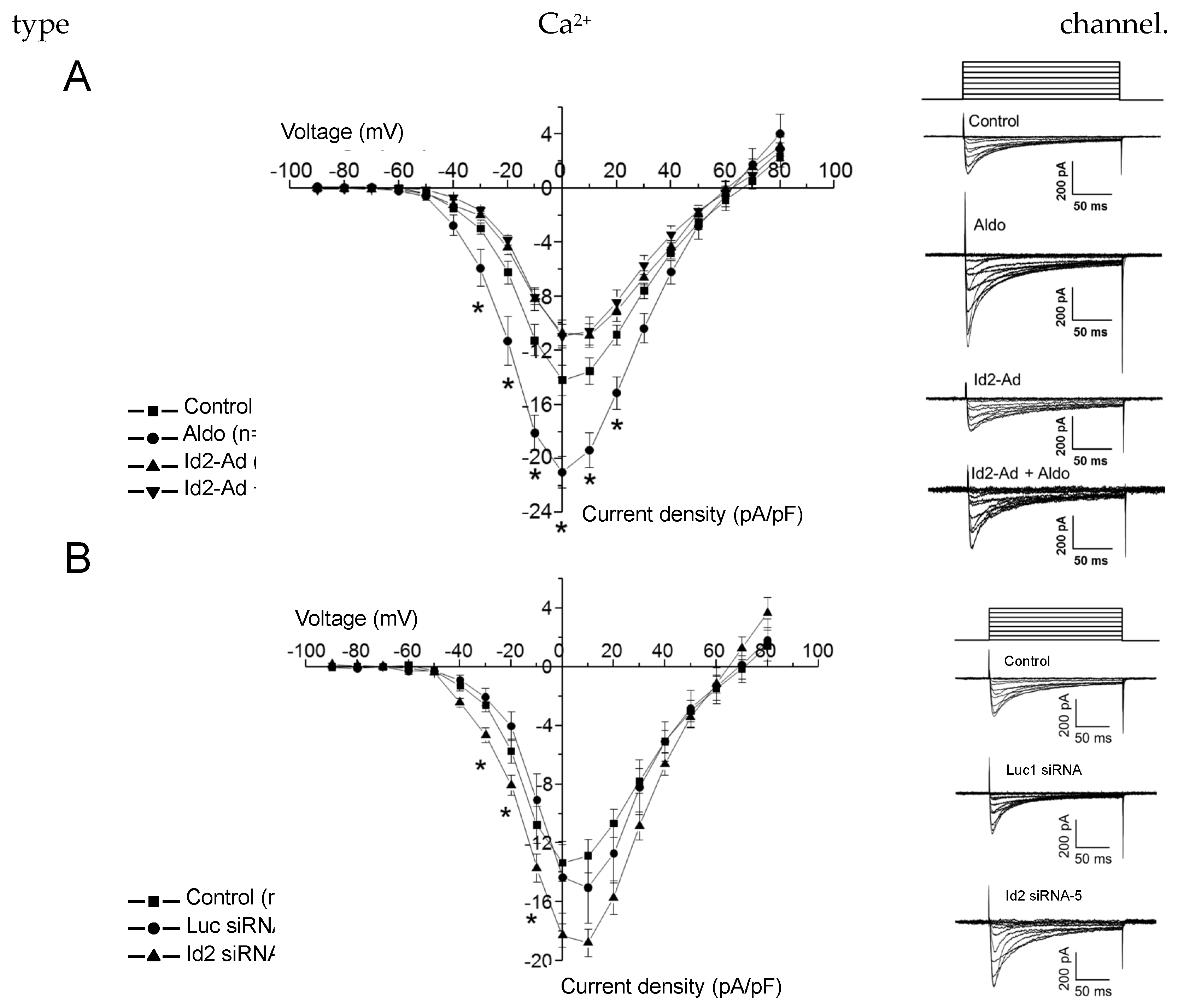
2.3. Id2 Expression Modulate the Voltage-Gated Calcium Currents in Cardiomyocytes
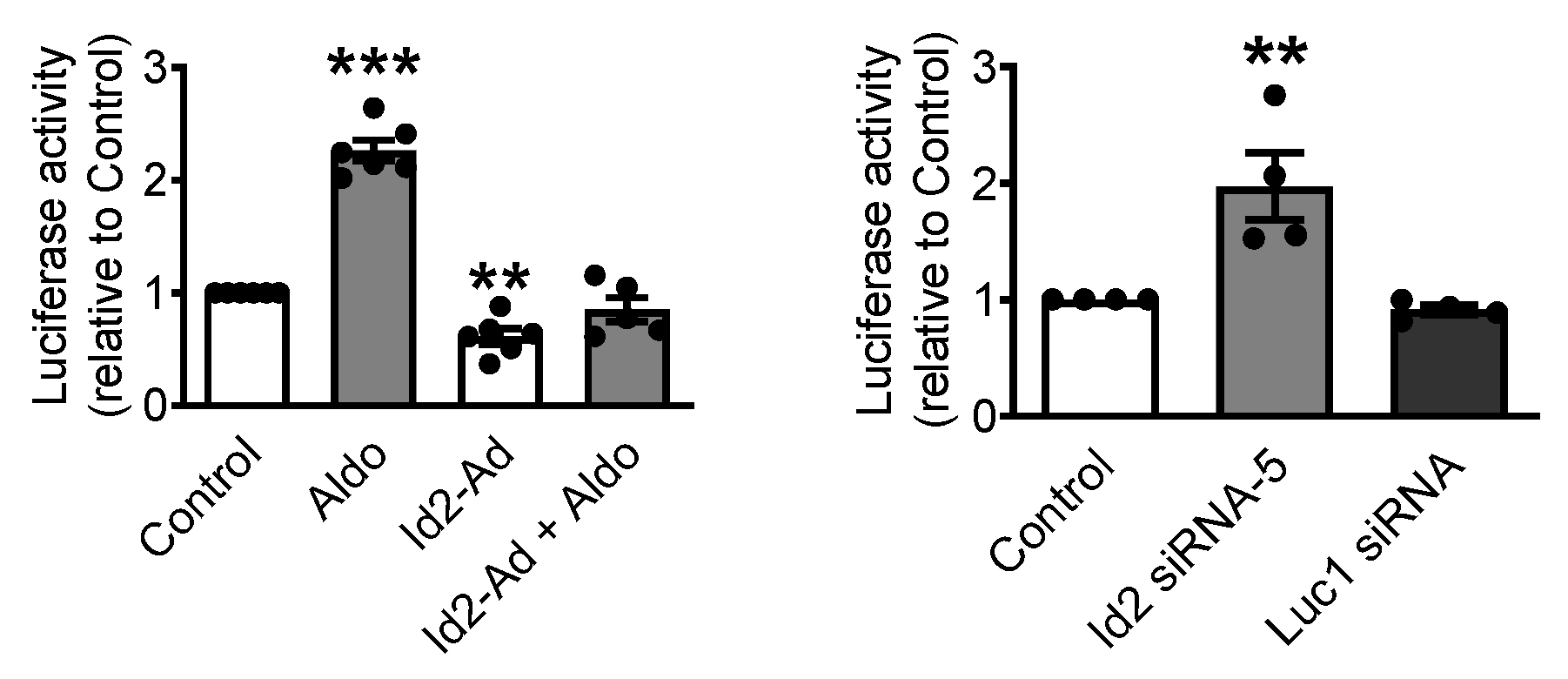
2.4. Id2 Represses CaV3.1 Expression at the Transcriptional Level
2.5. Id2 Prevents the Aldosterone-Stimulated Increase in Voltage-Gated Calcium Channels in Adult Mouse Heart
3. Material and Methods
3.1. Animals
3.2. Id2 Transgenic Mice
3.3. Aldosterone Treatment
3.4. Cardiomyocyte Isolation and Culture
3.5. RT-PCR Analysis
3.6. Real-Time Quantitative PCR Analysis
3.7. Luciferase Reporter Assay
3.8. RNA Interference
3.9. Plasmid Construction
3.10. Adenovirus
3.11. Western Blot
3.12. Electrophysiology
3.13. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Essick, E.E.; Sam, F. Cardiac hypertrophy and fibrosis in the metabolic syndrome: A role for aldosterone and the mineralocorticoid receptor. Int. J. Hypertens. 2011, 2011, 346985. [Google Scholar] [CrossRef] [Green Version]
- Lother, A.; Moser, M.; Bode, C.; Feldman, R.D.; Hein, L. Mineralocorticoids in the heart and vasculature: New insights for old hormones. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Funder, J.W. Aldosterone, hypertension and heart failure: Insights from clinical trials. Hypertens. Res. 2010, 33, 872–875. [Google Scholar] [CrossRef] [Green Version]
- Gravez, B.; Tarjus, A.; Jaisser, F. Mineralocorticoid receptor and cardiac arrhythmia. Clin. Exp. Pharmacol. Physiol. 2013, 40, 910–915. [Google Scholar] [CrossRef]
- Patramela, L.; Concistrè, A.; Olmati, F.; Saracino, V.; Chimenti, C.; Frustaci, A.; Russo, M.A.; Letizia, C. Cardiomyopathies and Adrenal Diseases. Int. J. Mol. Sci. 2020, 21, 5047. [Google Scholar] [CrossRef]
- Funder, J.W. Aldosterone and mineralocorticoid receptors: A personal reflection. Mol. Cell. Endocrinol. 2021, 350, 146–150. [Google Scholar] [CrossRef]
- Benitah, J.P.; Vassort, G. Aldosterone upregulates Ca(2+) current in adult rat cardiomyocytes. Circ. Res. 1999, 85, 1139–1145. [Google Scholar] [CrossRef] [Green Version]
- Ouvrard-Pascaud, A.; Sainte-Marie, Y.; Benitah, J.P.; Perrier, R.; Soukaseum, C.; Nguyen Dinh Cat, A.; Royer, A.; Le Quang, K.; Charpentier, F.; Demolombe, S. Conditional mineralocorticoid receptor expression in the heart leads to life-threatening arrhythmias. Circulation 2005, 111, 3025–3033. [Google Scholar] [CrossRef] [Green Version]
- Martin-Fernandez, B.; Miana, M.; De Las Heras, N.; Ruiz-Hurtado, G.; Fernandez-Velasco, M.; Bas, M.; Ballesteros, S.; Lahera, V.; Cachofeiro, V.; Delgado, C. Cardiac L-type calcium current is increased in a model of hyperaldosteronism in the rat. Exp. Physiol. 2009, 94, 675–683. [Google Scholar] [CrossRef]
- Maturana, A.; Lenglet, S.; Python, M.; Kuroda, S.; Rossier, M.F. Role of the T-Type Calcium Channel Ca(V)3.2 in the Chronotropic Action of Corticosteroids in Isolated Rat Ventricular Myocytes. Endocrinology 2009, 150, 3726–3734. [Google Scholar] [CrossRef]
- Lalevee, N.; Rebsamen, M.C.; Barrere-Lemaire, S.; Perrier, E.; Nargeot, J.; Benitah, J.P.; Rossier, M.F. Aldosterone increases T-type calcium channel expression and in vitro beating frequency in neonatal rat cardiomyocytes. Cardiovasc. Res. 2005, 67, 216–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferron, L.; Ruchon, Y.; Renaud, J.F.; Capuano, V. T-type Ca2+ signalling regulates aldosterone-induced CREB activation and cell death through PP2A activation in neonatal cardiomyocytes. Cardiovasc. Res. 2011, 90, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, K.; Iijima, T. Cardiac T-type Ca(2+) channels in the heart. J. Mol. Cell. Cardiol. 2010, 48, 65–70. [Google Scholar] [CrossRef]
- Chiang, C.S.; Huang, C.H.; Chieng, H.L.; Chang, Y.T.; Chang, D.R.; Chen, J.J.; Chen, Y.C.; Chen, Y.H.; Shin, H.S.; Campbell, K.P.; et al. The Ca(v)3.2 T-Type Ca2+ Channel Is Required for Pressure Overload-Induced Cardiac Hypertrophy in Mice. Circ. Res. 2009, 104, 522–530. [Google Scholar] [CrossRef] [Green Version]
- BenMohamed, F.; Ferron, L.; Ruchon, Y.; Gouadon, E.; Renaud, J.F.; Capuano, V. Regulation of T-type Cav3.1 channels expression by synthetic glucocorticoid dexamethasone in neonatal cardiac myocytes. Mol. Cell. Biochem. 2010, 335, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Koyama, R.; Mannic, T.; Ito, J.; Amar, L.; Zennaro, M.-C.; Rossier, M.F.; Maturana, A.D. MicroRNA204 is necessary for aldosterone-stimulated T-type calcium channel expression in cardiomyocytes. Int. J. Mol. Sci. 2018, 19, 2941. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.V.; Jiang, S.; Palasingam, P.; Kolatkar, P.R. A divalent ion is crucial in the structure and dominant-negative function of ID proteins, a class of helix-loop-helix transcription regulators. PLoS ONE 2012, 7, e48591. [Google Scholar] [CrossRef]
- Fraidenraich, D.; Stillwell, E.; Romero, E.; Wilkes, D.; Manova, K.; Basson, C.T.; Benezra, R. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science 2004, 306, 247–252. [Google Scholar] [CrossRef] [Green Version]
- Moskowitz, I.P.; Wang, J.; Peterson, M.A.; Pu, W.T.; Mackinnon, A.C.; Oxburgh, L.; Chu, G.C.; Sarkar, M.; Berul, C.; Smoot, L.; et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proc. Natl. Acad. Sci. USA 2011, 108, 4006–4011. [Google Scholar] [CrossRef] [Green Version]
- Moskowitz, I.P.; Kim, J.B.; Moore, M.L.; Wolf, C.M.; Peterson, M.A.; Shendure, J.; Nobrega, M.A.; Yokota, Y.; Berul, C.; Izumo, S.; et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell 2007, 129, 1365–1376. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Liu, C.J.; Huang, Y.; Yu, J.; Kong, W.; Lengyel, P. p204 protein overcomes the inhibition of the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes by Id proteins. J. Biol. Chem. 2006, 281, 14893–14906. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.Y.; Kim, W.H.; Kim, J.; Park, S.I. Induction of Id2 expression by cardiac transcription factors GATA4 and Nkx2. J. Cell. Biochem. 2008, 103, 182–914. [Google Scholar] [CrossRef]
- Messaoudi, S.; Gravez, B.; Tarjus, A.; Pelloux, V.; Ouvrard-Pascaud, A.; Delcayre, C.; Samuel, J.; Launay, J.M.; Sierra-Ramos, C.; de la Rosa, D.A.; et al. Aldosteron-specific activation of cardiomyocyte mineralocorticoid receptor in-vivo. Hypertension 2013, 61, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jongbloed, M.R.; Vicente-Steijn, R.; Douglas, Y.L.; Wisse, L.J.; Mori, K.; Yokota, Y.; Bartelings, M.M.; Schalij, M.J.; Mahtab, E.A.; Poelmann, R.E.; et al. Expression of Id2 in the second heart field and cardiac defects in Id2 knock-out mice. Dev. Dyn. 2011, 240, 2561–2577. [Google Scholar] [CrossRef] [PubMed]
- Rossier, M.F.; Python, M.; Maturana, A.D. Contribution of Mineralocorticoid and Glucocorticoid Receptors to the Chronotropic and Hypertrophic Actions of Aldosterone in Neonatal Rat Ventricular Myocytes. Endocrinology 2010, 151, 2777–2787. [Google Scholar] [CrossRef] [Green Version]
- Perrier, E.; Kerfant, B.G.; Lalevee, N.; Bideaux, P.; Rossier, M.F.; Richard, S.; Gómez, A.M.; Benitah, J.P. Mineralocorticoid receptor antagonism prevents the electrical remodeling that precedes cellular hypertrophy after myocardial infarction. Circulation 2004, 110, 776–783. [Google Scholar] [CrossRef] [Green Version]
- Bienvenu, L.A.; Reichelt, M.E.; Delbridge, L.M.D.; Young, M.J. Mineralocorticoid receptors and the heart, multiple cell types and multiple mechanisms: A focus on the cardiomyocyte. Clin. Sci. 2013, 125, 409–421. [Google Scholar] [CrossRef] [Green Version]
- Maturana, A.D.; Walchli, S.; Iwata, M.; Ryser, S.; Van Lint, J.; Hoshijima, M.; Schlegel, W.; Ikeda, Y.; Tanizawa, K.; Kuroda, S. Enigma homolog 1 scaffolds protein kinase D1 to regulate the activity of the cardiac L-type voltage-gated calcium channel. Cardiovasc. Res. 2008, 78, 458–465. [Google Scholar] [CrossRef] [Green Version]
- Kuwahara, K.; Saito, Y.; Takano, M.; Arai, Y.; Yasuno, S.; Nakagawa, Y.; Takahashi, N.; Adachi, Y.; Takemura, G.; Horie, M.; et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003, 22, 6310–6321. [Google Scholar] [CrossRef]
- Guo, A.; Wang, Y.; Chen, B.; Wang, Y.; Yuan, J.; Zhang, L.; Hall, D.; Wu, J.; Shi, Y.; Zhu, Q.; et al. E-C coupling structural protein junctophilin-2 encodes a stress-adaptative transcription regulator. Science 2018, 362, eaan3303. [Google Scholar] [CrossRef]


| WT (n = 5) | Id2 Tg B (n = 5) | Id2 Tg C (n = 5) | |
|---|---|---|---|
| Body weight (g) | 21.8 ± 1.8 | 21.1 ± 1.3 | 21.0 ± 0.5 |
| Whole heart weight (mg) | 120.9 ± 8.4 | 119.2 ± 8.5 | 117.4 ± 7.6 |
| Lung weight (mg) | 109.1 ± 4.3 | 108.8 ± 2.9 | 111.6 ± 2.8 |
| Liver weight (mg) | 1004.2 ± 13.6 | 973.3 ± 69.8 | 1014.2 ± 37.3 |
| WHW/BW | 5.54 ± 0.39 | 5.64 ± 0.15 | 5.58 ± 0.29 |
| WT Saline (n = 8) | Id2 Saline (n = 6) | WT Aldo (n = 8) | Id2 Aldo (n = 5) | ||
|---|---|---|---|---|---|
| Body weight (g) | 24.3 ± 1.2 | 23.8 ± 0.8 | 25.1 ± 0.4 | 24.3 ± 1.1 | |
| Whole heart weight (mg) | 127.5 ± 13.2 | 119.1 ± 9.6 | 155.8 ± 15.8 | 133.9 ± 5.8 | |
| Lung weight (mg) | 121.3 ± 10.3 | 111.9 ± 3.6 | 137.9 ± 9.6 | 126.7 ± 4.0 | |
| Liver weight (mg) | 1129.9 ± 77.4 | 1076.3 ± 63.3 | 1105.6 ± 37.9 | 1112.1 ± 45.3 | |
| WHW/BW | 5.22 ± 0.43 | 4.97 ± 0.28 | 6.20 ± 0.61 | 5.51 ± 0.11 | |
| Saline (n = 5) | Aldo (n = 8) | p Value | ||
|---|---|---|---|---|
| Body weight (g) | 23.8 ± 1.2 | 25.2 ± 0.8 | 0.318 | |
| Whole heart weight (mg) | 118.4 ± 14.2 | 149.8 ± 17.9 | 0.244 | |
| Lung weight (mg) | 116.1 ± 9.2 | 140.0 ± 10.2 | 0.138 | |
| Liver weight (mg) | 1081.6 ± 93.7 | 1141.6 ± 71.8 | 0.618 | |
| WHW/BW | 4.95 ± 0.46 | 5.96 ± 0.70 | 0.319 | |
| Forward 5’-3’ | Reverse 5’-3’ | |
|---|---|---|
| Rat Id2 | CTCCAAGCTCAAGGAACTGG | ATGCTGATGTCCGTGTTCAG |
| Rat CaV1.2 | AGCAACTTCCCTCAGACGTTTG | GCTTCTCATGGGACGGTGAT |
| Rat CaV3.1 | ACGCTGAGTCTCTCTGGTTTGTC | TGCTTACGTGGGACTTTTCAGA |
| Rat CaV3.2 | GGCGAAGAAGGCAAAGATGA | GCGTGACACTGGGCATGTT |
| Rat GAPDH | CAACTCCCTCAAGATTGTCAGCAA | GGCATGGACTGTGGTCATGA |
| Rat Hcn2 | GGACACTTTCTTCCTCATGGAC | CTCCGTGTTGTCCTCAATAAC |
| Rat Hcn4 | CTTCCTCATTGACTTGGTCCTC | TACTTCATCTTGATCCGCTGTG |
| Rat Kcnh2 | ACCTTCAACCTTCGAGATACCAAC | GCTCTGTGTCCTTGTCAGTACG |
| Rat Kcnd3 | GACAAGAACAAGCGGCAAGA | GCATTCATAGCGTGGGTAGT |
| Rat Scn5a | ACCTGCCTCTGAACTACACCATC | CCTTGGTCCAGTACAACTCTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, J.; Minemura, T.; Wälchli, S.; Niimi, T.; Fujihara, Y.; Kuroda, S.; Takimoto, K.; Maturana, A.D. Id2 Represses Aldosterone-Stimulated Cardiac T-Type Calcium Channels Expression. Int. J. Mol. Sci. 2021, 22, 3561. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073561
Ito J, Minemura T, Wälchli S, Niimi T, Fujihara Y, Kuroda S, Takimoto K, Maturana AD. Id2 Represses Aldosterone-Stimulated Cardiac T-Type Calcium Channels Expression. International Journal of Molecular Sciences. 2021; 22(7):3561. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073561
Chicago/Turabian StyleIto, Jumpei, Tomomi Minemura, Sébastien Wälchli, Tomoaki Niimi, Yoshitaka Fujihara, Shun’ichi Kuroda, Koichi Takimoto, and Andrés D. Maturana. 2021. "Id2 Represses Aldosterone-Stimulated Cardiac T-Type Calcium Channels Expression" International Journal of Molecular Sciences 22, no. 7: 3561. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073561







