Polypurine Reverse-Hoogsteen Hairpins as a Tool for Exon Skipping at the Genomic Level in Mammalian Cells
Abstract
:1. Introduction
2. Results
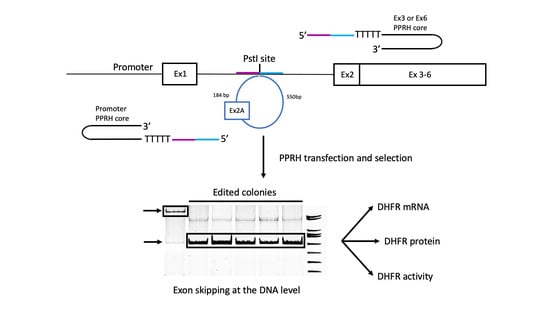
2.1. Design of Editing PPRHs
2.2. DNA Sequencing in Edited Colonies
2.3. DHFR mRNA Expression in Edited Cell Colonies
2.4. DHFR Protein Levels and Enzymatic Activity in Edited Cell Colonies
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Cell Culture
4.3. Transfection of PPRHs and Selection
4.4. DNA Sequencing
4.5. RNA Extraction and mRNA Analyses
4.6. Western Blot Analyses
4.7. DHFR Activity
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffman, E.P.; Brown, R.H.; Kunkel, L.M. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef]
- Mendell, J.R.; Shilling, C.; Leslie, N.D.; Flanigan, K.M.; Al-Dahhak, R.; Gastier-Foster, J.; Kneile, K.; Dunn, D.M.; Duval, B.; Aoyagi, A.; et al. Evidence-based path to newborn screening for duchenne muscular dystrophy. Ann. Neurol. 2012, 71, 304–314. [Google Scholar] [CrossRef]
- Koenig, M.; Beggs, A.H.; Moyer, M.; Scherpf, S.; Heindrich, K.; Bettecken, T.; Meng, G.; Müller, C.R.; Lindlöf, M.; Kaariainen, H.; et al. The molecular basis for duchenne versus becker muscular dystrophy: Correlation of severity with type of deletion. Am. J. Hum. Genet. 1989, 45, 498–506. [Google Scholar]
- Aartsma-Rus, A.; Fokkema, I.; Verschuuren, J.; Ginjaar, I.; Van Deutekom, J.; Van Ommen, G.J.; Den Dunnen, J.T. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 2009, 30, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Straub, V.; Hemmings, R.; Haas, M.; Schlosser-Weber, G.; Stoyanova-Beninska, V.; Mercuri, E.; Muntoni, F.; Sepodes, B.; Vroom, E.; et al. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther. 2017, 25, 251–259. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Garn, H.; Unger, S.D.; Renz, H. Antisense molecules: A new class of drugs. J. Allergy Clin. Immunol. 2016, 137, 1334–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, A.G.L.; Wood, M.J.A. Splicing therapy for neuromuscular disease. Mol. Cell. Neurosci. 2013, 56, 169–185. [Google Scholar] [CrossRef]
- Sardone, V.; Zhou, H.; Muntoni, F.; Ferlini, A.; Falzarano, M.S. Antisense oligonucleotide-based therapy for neuromuscular disease. Molecules 2017, 22, 563. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, J.; Xiao, X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc. Natl. Acad. Sci. USA 2000, 97, 13714–13719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echigoya, Y.; Yokota, T. Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther. 2014, 24, 57–68. [Google Scholar] [CrossRef]
- Guncay, A.; Yokota, T. Antisense oligonucleotide drugs for Duchenne muscular dystrophy: How far have we come and what does the future hold? Future Med. Chem. 2015, 7, 1631–1635. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krieg, A.M. Exon skipping therapy for Duchenne muscular dystrophy. Adv. Drug Deliv. Rev. 2015, 87, 104–107. [Google Scholar] [CrossRef]
- Matos, L.; Vilela, R.; Rocha, M.; Santos, J.I.; Coutinho, M.F.; Gaspar, P.; Prata, M.J.; Alves, S. Development of an Antisense Oligonucleotide-Mediated Exon Skipping Therapeutic Strategy for Mucolipidosis II: Validation at RNA Level. Hum. Gene Ther. 2020, 31, 775–783. [Google Scholar] [CrossRef]
- Gramlich, M.; Pane, L.S.; Zhou, Q.; Chen, Z.; Murgia, M.; Schötterl, S.; Goedel, A.; Metzger, K.; Brade, T.; Parrotta, E.; et al. Antisense-mediated exon skipping: A therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 2015, 5, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Park, K.H.; Zhao, L.; Xu, J.; El Refaey, M.; Gao, Y.; Zhu, H.; Ma, J.; Han, R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2016, 24, 564–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, P.; Wu, F.; Mosenson, J.; Zhang, H.; He, T.C.; Wu, W.S. CRISPR/Cas9-Mediated Genome Editing Corrects Dystrophin Mutation in Skeletal Muscle Stem Cells in a Mouse Model of Muscle Dystrophy. Mol. Ther. Nucleic Acids 2017, 7, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [Green Version]
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef] [Green Version]
- Amoasii, L.; Hildyard, J.C.W.; Li, H.; Sanchez-Ortiz, E.; Mireault, A.; Caballero, D.; Harron, R.; Stathopoulou, T.R.; Massey, C.; Shelton, J.M.; et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018, 362, 86–91. [Google Scholar] [CrossRef] [Green Version]
- de Almagro, M.C.; Coma, S.; Noé, V.; Ciudad, C.J. Polypurine hairpins directed against the template strand of DNA knock down the expression of mammalian genes. J. Biol. Chem. 2009, 284, 11579–11589. [Google Scholar] [CrossRef] [Green Version]
- Coma, S.; Noé, V.; Eritja, R.; Ciudad, C.J. Strand displacement of double-stranded DNA by triplex-forming antiparallel purine-hairpins. Oligonucleotides 2005, 15, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Félix, A.J.; Ciudad, C.J.; Noé, V. Correction of the aprt Gene Using Repair-Polypurine Reverse Hoogsteen Hairpins in Mammalian Cells. Mol. Ther. Nucleic Acids 2020, 19, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Ciudad, C.J.; Rodríguez, L.; Villalobos, X.; Félix, A.J.; Noé, V. Polypurine Reverse Hoogsteen Hairpins as a Gene Silencing Tool for Cancer. Curr. Med. Chem. 2017, 24, 2809–2826. [Google Scholar] [CrossRef] [PubMed]
- Félix, A.J.; Solé, A.; Noé, V.; Ciudad, C.J. Gene Correction of Point Mutations Using PolyPurine Reverse Hoogsteen Hairpins Technology. Front. Genome Ed. 2020. [Google Scholar] [CrossRef]
- Chen, I.T.; Chasin, L.A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol. Cell. Biol. 1993, 13, 289–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solé, A.; Ciudad, C.J.; Chasin, L.A.; Noé, V. Correction of point mutations at the endogenous locus of the dihydrofolate reductase gene using repair-PolyPurine Reverse Hoogsteen hairpins in mammalian cells. Biochem. Pharmacol. 2016, 110–111, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Frazier, K.S. Antisense Oligonucleotide Therapies:The Promise and the Challenges from a Toxicologic Pathologist’s Perspective. Toxicol. Pathol. 2015, 43, 78–89. [Google Scholar] [CrossRef]
- Félix, A.J.; Ciudad, C.J.; Noé, V. Functional pharmacogenomics and toxicity of PolyPurine Reverse Hoogsteen hairpins directed against survivin in human cells. Biochem. Pharmacol. 2018, 155, 8–20. [Google Scholar] [CrossRef]
- Datta, H.J.; Chan, P.P.; Vasquez, K.M.; Gupta, R.C.; Glazer, P.M. Triplex-induced recombination in human cell-free extracts: Dependence on XPA and HsRad51. J. Biol. Chem. 2001, 276, 18018–18023. [Google Scholar] [CrossRef] [Green Version]
- Knauert, M.P.; Kalish, J.M.; Hegan, D.C.; Glazer, P.M. Triplex-Stimulated Intermolecular Recombination at a Single-Copy Genomic Target. Mol. Ther. 2006, 14, 392–400. [Google Scholar] [CrossRef]
- Solé, A.; Villalobos, X.; Ciudad, C.J.; Noé, V. Repair of single-point mutations by polypurine reverse hoogsteen hairpins. Hum. Gene Ther. Methods 2014, 25, 288–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggio, I.; Stefanucci, L.; Janssen, J.M.; Liu, J.; Chen, X.; Mouly, V.; Gonçalves, M.A.F.V. Selection-free gene repair after adenoviral vector transduction of designer nucleases: Rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016, 44, 1449–1470. [Google Scholar] [CrossRef] [Green Version]
- Allen, F.; Crepaldi, L.; Alsinet, C.; Strong, A.J.; Kleshchevnikov, V.; De Angeli, P.; Páleníková, P.; Khodak, A.; Kiselev, V.; Kosicki, M.; et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018. [Google Scholar] [CrossRef]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.R.; Haeussler, M.; Watanabe, C.; Janakiraman, V.; Lund, J.; Modrusan, Z.; Stinson, J.; Bei, Q.; Buechler, A.; Yu, C.; et al. CRISPR off-target analysis in genetically engineered rats and mice. Nat. Methods 2018, 15, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, K.A.; Wu, W.H.; Colgan, D.F.; Tsang, S.H.; Bassuk, A.G.; Mahajan, V.B. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods 2017, 14, 547–548. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef] [Green Version]
- Ciudad, C.J.; Urlaub, G.; Chasin, L.A. Deletion analysis of the Chinese hamster dihydrofolate reductase gene promoter. J. Biol. Chem. 1988, 263, 16274–16282. [Google Scholar] [CrossRef]







| Name | Editing PPRH Sequence | Location of the Target for the PPRH Core |
|---|---|---|
| LDSHpPrI1UPstI | 5’CTGCAGCCGGCGGGCCTACCTTTTTGGGGAAGGAAAAGTGGGTGACATTTTTACAGTGGGTGAAAAGGAAGGGG3’ | Promoter |
| LDSHpPrI1UDstI | 5’ACCGCGGGCATGGGCAAGGGCTGCAGCCGGCGGGCCTACCTTTTTGGGGAAGGAAAAGTGGGTGACATTTTTACAGTGGGTGAAAAGGAAGGGG3’ | Promoter |
| LDSHpE6I1DPstI | 5’CCCTTGCCCATGCCCGCGGTTTTTTAGGAGGAAAAAGGCATCAAGTTTTTGAACTACGGAAAAAGGAGGA3’ | Exon 6 |
| LDSHpE6I1UDPstI | 5’GGTAGGCCCGCCGGCTGCAGCCCTTGCCCATGCCCGCGGTTTTTTAGGAGGAAAAAGGCATCAAGTTTTTGAACTACGGAAAAAGGAGGA3’ | Exon 6 |
| LDSHpE3I1UDPstI | 5’GGTAGGCCCGCCGGCTGCAGCCCTTGCCCATGCCCGCGGTTTTTTGGACCAAGAGGTAAGGATTTTTAGGAATGGAGAACCAGG3’ | Exon 3 |
| UDPst1 | 5’GGTAGGCCCGCCGGCTGCAGCCCTTGCCCATGCCCGCGGT3’ | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noé, V.; Ciudad, C.J. Polypurine Reverse-Hoogsteen Hairpins as a Tool for Exon Skipping at the Genomic Level in Mammalian Cells. Int. J. Mol. Sci. 2021, 22, 3784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073784
Noé V, Ciudad CJ. Polypurine Reverse-Hoogsteen Hairpins as a Tool for Exon Skipping at the Genomic Level in Mammalian Cells. International Journal of Molecular Sciences. 2021; 22(7):3784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073784
Chicago/Turabian StyleNoé, Véronique, and Carlos J. Ciudad. 2021. "Polypurine Reverse-Hoogsteen Hairpins as a Tool for Exon Skipping at the Genomic Level in Mammalian Cells" International Journal of Molecular Sciences 22, no. 7: 3784. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22073784