A Versatile Toolkit for Semi-Automated Production of Fluorescent Chemokines to Study CCR7 Expression and Functions
Abstract
:1. Introduction
2. Results
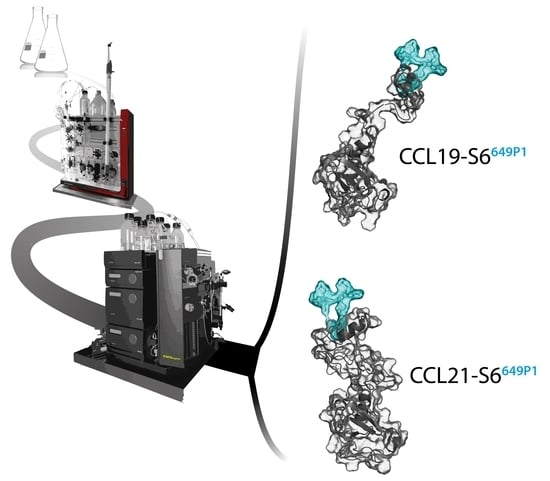
2.1. Recombinant Production, Automated Chromatography Purification and Enzymatic Fluorescent Labelling of CCL19 and CCL21
2.2. Fluorescent CCL19-S6649P1 and CCL21-S6649P1 Are Biologically Active and Efficiently Elicit Early CCR7 Signal Transduction Pathways
2.3. CCL19-S6649P1 Specifically Binds to and Interacts with CCR7 on Transfected Cells
2.4. CCL19-S6649P1 Is Internalized by CCR7-Expressing Cells
2.5. CCL19-S6649P1 Is a Versatile Tool to Stain CCR7-Expressing Primary Lymphocyte Subsets
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Plasmids
4.3. Chemokine Production and Fluorescent Labelling
4.4. SDS-PAGE and Western Blot Analysis
4.5. Cell Lines and Transfection
4.6. Isolation of Primary Human and Mouse Leukocytes
4.7. Chemokine-Mediated [Ca2+]i-Mobilization
4.8. Chemotaxis Assay
4.9. Bioluminescence Resonance Energy Transfer (BRET)
4.10. Chemokine Binding and Uptake Assay
4.11. Chemokine Receptor Surface Staining
4.12. Confocal Live Cell Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Legler, D.F.; Thelen, M. Chemokines: Chemistry, Biochemistry and Biological Function. Chimia 2016, 70, 856–859. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [Green Version]
- Nagira, M.; Imai, T.; Hieshima, K.; Kusuda, J.; Ridanpaa, M.; Takagi, S.; Nishimura, M.; Kakizaki, M.; Nomiyama, H.; Yoshie, O. Molecular cloning of a novel human CC chemokine secondary lymphoid-tissue chemokine that is a potent chemoattractant for lymphocytes and mapped to chromosome 9p13. J. Biol. Chem. 1997, 272, 19518–19524. [Google Scholar] [CrossRef] [Green Version]
- Hirose, J.; Kawashima, H.; Swope Willis, M.; Springer, T.A.; Hasegawa, H.; Yoshie, O.; Miyasaka, M. Chondroitin sulfate B exerts its inhibitory effect on secondary lymphoid tissue chemokine (SLC) by binding to the C-terminus of SLC. Biochim. Biophys. Acta 2002, 1571, 219–224. [Google Scholar] [CrossRef]
- Veldkamp, C.T.; Kiermaier, E.; Gabel-Eissens, S.J.; Gillitzer, M.L.; Lippner, D.R.; DiSilvio, F.A.; Mueller, C.J.; Wantuch, P.L.; Chaffee, G.R.; Famiglietti, M.W.; et al. Solution structure of CCL19 and identification of overlapping CCR7 and PSGL-1 binding sites. Biochemistry 2015, 54, 4163–4166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, K.; Lammermann, T.; Bruckner, M.; Legler, D.F.; Polleux, J.; Spatz, J.P.; Schuler, G.; Forster, R.; Lutz, M.B.; Sorokin, L.; et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 2010, 32, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Hauser, M.A.; Legler, D.F. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J. Leukoc. Biol. 2016, 99, 869–882. [Google Scholar] [CrossRef] [Green Version]
- Luther, S.A.; Tang, H.L.; Hyman, P.L.; Farr, A.G.; Cyster, J.G. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc. Natl. Acad. Sci. USA 2000, 97, 12694–12699. [Google Scholar] [CrossRef] [Green Version]
- Carlsen, H.S.; Haraldsen, G.; Brandtzaeg, P.; Baekkevold, E.S. Disparate lymphoid chemokine expression in mice and men: No evidence of CCL21 synthesis by human high endothelial venules. Blood 2005, 106, 444–446. [Google Scholar] [CrossRef] [Green Version]
- Forster, R.; Davalos-Misslitz, A.C.; Rot, A. CCR7 and its ligands: Balancing immunity and tolerance. Nat. Rev. Immunol. 2008, 8, 362–371. [Google Scholar] [CrossRef]
- Bruckner, M.; Dickel, D.; Singer, E.; Legler, D.F. Distinct modulation of chemokine expression patterns in human monocyte-derived dendritic cells by prostaglandin E(2). Cell Immunol. 2012, 276, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Weber, M.; Hauschild, R.; Schwarz, J.; Moussion, C.; de Vries, I.; Legler, D.F.; Luther, S.A.; Bollenbach, T.; Sixt, M. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 2013, 339, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Russo, E.; Teijeira, A.; Vaahtomeri, K.; Willrodt, A.H.; Bloch, J.S.; Nitschke, M.; Santambrogio, L.; Kerjaschki, D.; Sixt, M.; Halin, C. Intralymphatic CCL21 Promotes Tissue Egress of Dendritic Cells through Afferent Lymphatic Vessels. Cell Rep. 2016, 14, 1723–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, K.D.; Legler, D.F.; Purvanov, V.; Ohigashi, I.; Takahama, Y.; Parnell, S.M.; White, A.J.; Jenkinson, W.E.; Anderson, G. Medullary stromal cells synergize their production and capture of CCL21 for T-cell emigration from neonatal mouse thymus. Blood Adv. 2021, 5, 99–112. [Google Scholar] [CrossRef]
- Willimann, K.; Legler, D.F.; Loetscher, M.; Roos, R.S.; Delgado, M.B.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. The chemokine SLC is expressed in T cell areas of lymph nodes and mucosal lymphoid tissues and attracts activated T cells via CCR7. Eur. J. Immunol. 1998, 28, 2025–2034. [Google Scholar] [CrossRef]
- Yoshida, R.; Nagira, M.; Kitaura, M.; Imagawa, N.; Imai, T.; Yoshie, O. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J. Biol. Chem. 1998, 273, 7118–7122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallusto, F.; Lenig, D.; Forster, R.; Lipp, M.; Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Legler, D.F.; Uetz-von Allmen, E.; Hauser, M.A. CCR7: Roles in cancer cell dissemination, migration and metastasis formation. Int. J. Biochem. Cell Biol. 2014, 54, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Legler, D.F.; Thelen, M. New insights in chemokine signaling. F1000Research 2018, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Nibbs, R.J.; Graham, G.J. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 2013, 13, 815–829. [Google Scholar] [CrossRef]
- Kohout, T.A.; Nicholas, S.L.; Perry, S.J.; Reinhart, G.; Junger, S.; Struthers, R.S. Differential Desensitization, Receptor Phosphorylation, beta-Arrestin Recruitment, and ERK1/2 Activation by the Two Endogenous Ligands for the CC Chemokine Receptor 7. J. Biol. Chem. 2004, 279, 23214–23222. [Google Scholar] [CrossRef] [Green Version]
- Otero, C.; Eisele, P.S.; Schaeuble, K.; Groettrup, M.; Legler, D.F. Distinct motifs in the chemokine receptor CCR7 regulate signal transduction, receptor trafficking and chemotaxis. J. Cell Sci. 2008, 121, 2759–2767. [Google Scholar] [CrossRef] [Green Version]
- Schaeuble, K.; Hauser, M.A.; Singer, E.; Groettrup, M.; Legler, D.F. Cross-talk between TCR and CCR7 signaling sets a temporal threshold for enhanced T lymphocyte migration. J. Immunol. 2011, 187, 5645–5652. [Google Scholar] [CrossRef] [Green Version]
- Byers, M.A.; Calloway, P.A.; Shannon, L.; Cunningham, H.D.; Smith, S.; Li, F.; Fassold, B.C.; Vines, C.M. Arrestin 3 mediates endocytosis of CCR7 following ligation of CCL19 but not CCL21. J. Immunol. 2008, 181, 4723–4732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zidar, D.A.; Violin, J.D.; Whalen, E.J.; Lefkowitz, R.J. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc. Natl. Acad. Sci. USA 2009, 106, 9649–9654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laufer, J.M.; Hauser, M.A.; Kindinger, I.; Purvanov, V.; Pauli, A.; Legler, D.F. Chemokine Receptor CCR7 Triggers an Endomembrane Signaling Complex for Spatial Rac Activation. Cell Rep. 2019, 29, 995–1009. [Google Scholar] [CrossRef] [Green Version]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [Green Version]
- Hauser, M.A.; Kindinger, I.; Laufer, J.M.; Spate, A.K.; Bucher, D.; Vanes, S.L.; Krueger, W.A.; Wittmann, V.; Legler, D.F. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J. Leukoc. Biol. 2016, 99, 993–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, M.A.; Schaeuble, K.; Kindinger, I.; Impellizzieri, D.; Krueger, W.A.; Hauck, C.R.; Boyman, O.; Legler, D.F. Inflammation-Induced CCR7 Oligomers Form Scaffolds to Integrate Distinct Signaling Pathways for Efficient Cell Migration. Immunity 2016, 44, 59–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, L.B.; Cerovic, V.; Milling, S.W.; Graham, G.J.; Hansell, C.A.; Nibbs, R.J. Characterization of conventional and atypical receptors for the chemokine CCL2 on mouse leukocytes. J. Immunol. 2014, 193, 400–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansell, C.A.H.; Love, S.; Pingen, M.; Wilson, G.J.; MacLeod, M.; Graham, G.J. Analysis of lung stromal expression of the atypical chemokine receptor ACKR2 reveals unanticipated expression in murine blood endothelial cells. Eur. J. Immunol. 2020, 50, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Purvanov, V.; Matti, C.; Samson, G.P.B.; Kindinger, I.; Legler, D.F. Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. Int. J. Mol. Sci. 2018, 19, 3876. [Google Scholar] [CrossRef] [Green Version]
- Veldkamp, C.T.; Koplinski, C.A.; Jensen, D.R.; Peterson, F.C.; Smits, K.M.; Smith, B.L.; Johnson, S.K.; Lettieri, C.; Buchholz, W.G.; Solheim, J.C.; et al. Production of Recombinant Chemokines and Validation of Refolding. Methods Enzymol. 2016, 570, 539–565. [Google Scholar]
- Kawamura, T.; Stephens, B.; Qin, L.; Yin, X.; Dores, M.R.; Smith, T.H.; Grimsey, N.; Abagyan, R.; Trejo, J.; Kufareva, I.; et al. A general method for site specific fluorescent labeling of recombinant chemokines. PLoS ONE 2014, 9, e81454. [Google Scholar] [CrossRef] [Green Version]
- Mofid, M.R.; Finking, R.; Marahiel, M.A. Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4’-phosphopantetheinyl transferases AcpS and Sfp. J. Biol. Chem. 2002, 277, 17023–17031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehring, A.M.; Lambalot, R.H.; Vogel, K.W.; Drueckhammer, D.G.; Walsh, C.T. Ability of Streptomyces spp. acyl carrier proteins and coenzyme A analogs to serve as substrates in vitro for E. coli holo-ACP synthase. Chem. Biol. 1997, 4, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Flugel, R.S.; Hwangbo, Y.; Lambalot, R.H.; Cronan, J.E., Jr.; Walsh, C.T. Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J. Biol. Chem. 2000, 275, 959–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Lin, A.J.; Golan, D.E.; Walsh, C.T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006, 1, 280–285. [Google Scholar] [CrossRef]
- Zhou, Z.; Cironi, P.; Lin, A.J.; Xu, Y.; Hrvatin, S.; Golan, D.E.; Silver, P.A.; Walsh, C.T.; Yin, J. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem. Biol. 2007, 2, 337–346. [Google Scholar] [CrossRef]
- Clark-Lewis, I.; Kim, K.S.; Rajarathnam, K.; Gong, J.H.; Dewald, B.; Moser, B.; Baggiolini, M.; Sykes, B.D. Structure-activity relationships of chemokines. J. Leukoc. Biol. 1995, 57, 703–711. [Google Scholar] [CrossRef]
- Moepps, B.; Thelen, M. Monitoring Scavenging Activity of Chemokine Receptors. Methods Enzymol. 2016, 570, 87–118. [Google Scholar] [PubMed]
- Matti, C.; D’Uonnolo, G.; Artinger, M.; Melgrati, S.; Salnikov, A.; Thelen, S.; Purvanov, V.; Strobel, T.D.; Spannagel, L.; Thelen, M.; et al. CCL20 is a novel ligand for the scavenging atypical chemokine receptor 4. J. Leukoc. Biol. 2020, 107, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Otero, C.; Groettrup, M.; Legler, D.F. Opposite Fate of Endocytosed CCR7 and Its Ligands: Recycling versus Degradation. J. Immunol. 2006, 177, 2314–2323. [Google Scholar] [CrossRef] [Green Version]
- Barmore, A.J.; Castex, S.M.; Gouletas, B.A.; Griffith, A.J.; Metz, S.W.; Muelder, N.G.; Populin, M.J.; Sackett, D.M.; Schuster, A.M.; Veldkamp, C.T. Transferring the C-terminus of the chemokine CCL21 to CCL19 confers enhanced heparin binding. Biochem. Biophys. Res. Commun. 2016, 477, 602–606. [Google Scholar] [CrossRef] [Green Version]
- De Paz, J.L.; Moseman, E.A.; Noti, C.; Polito, L.; von Andrian, U.H.; Seeberger, P.H. Profiling heparin-chemokine interactions using synthetic tools. ACS Chem. Biol. 2007, 2, 735–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardi, G.; Lipp, M.; Baggiolini, M.; Loetscher, P. The T cell chemokine receptor CCR7 is internalized on stimulation with ELC, but not with SLC. Eur. J. Immunol. 2001, 31, 3291–3297. [Google Scholar] [CrossRef]
- Lu, Q.; Burns, M.C.; McDevitt, P.J.; Graham, T.L.; Sukman, A.J.; Fornwald, J.A.; Tang, X.; Gallagher, K.T.; Hunsberger, G.E.; Foley, J.J.; et al. Optimized procedures for producing biologically active chemokines. Protein Expr. Purif. 2009, 65, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Peterson, F.C.; Hayes, P.L.; Mattmiller, J.E.; Haugner, J.C., 3rd; de la Cruz, N.; Volkman, B.F. On-column refolding of recombinant chemokines for NMR studies and biological assays. Protein Expr. Purif. 2007, 52, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpe, S.; Cameroni, E.; Moepps, B.; Thelen, S.; Apuzzo, T.; Thelen, M. CCR2 Acts as Scavenger for CCL2 during Monocyte Chemotaxis. PLoS ONE 2012, 7, e37208. [Google Scholar] [CrossRef] [PubMed]
- Ameti, R.; Melgrati, S.; Radice, E.; Cameroni, E.; Hub, E.; Thelen, S.; Rot, A.; Thelen, M. Characterization of a chimeric chemokine as a specific ligand for ACKR3. J. Leukoc. Biol. 2018, 104, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Radice, E.; Ameti, R.; Melgrati, S.; Foglierini, M.; Antonello, P.; Stahl, R.A.K.; Thelen, S.; Jarrossay, D.; Thelen, M. Marginal Zone Formation Requires ACKR3 Expression on B Cells. Cell Rep. 2020, 32, 107951. [Google Scholar] [CrossRef]
- Matti, C.; Salnikov, A.; Artinger, M.; D’Agostino, G.; Kindinger, I.; Uguccioni, M.; Thelen, M.; Legler, D.F. ACKR4 Recruits GRK3 Prior to β-Arrestins but Can Scavenge Chemokines in the Absence of β-Arrestins. Front. Immunol. 2020, 11, 720. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artinger, M.; Matti, C.; Gerken, O.J.; Veldkamp, C.T.; Legler, D.F. A Versatile Toolkit for Semi-Automated Production of Fluorescent Chemokines to Study CCR7 Expression and Functions. Int. J. Mol. Sci. 2021, 22, 4158. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22084158
Artinger M, Matti C, Gerken OJ, Veldkamp CT, Legler DF. A Versatile Toolkit for Semi-Automated Production of Fluorescent Chemokines to Study CCR7 Expression and Functions. International Journal of Molecular Sciences. 2021; 22(8):4158. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22084158
Chicago/Turabian StyleArtinger, Marc, Christoph Matti, Oliver J. Gerken, Christopher T. Veldkamp, and Daniel F. Legler. 2021. "A Versatile Toolkit for Semi-Automated Production of Fluorescent Chemokines to Study CCR7 Expression and Functions" International Journal of Molecular Sciences 22, no. 8: 4158. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22084158