A Tryptophan-Deficient Diet Induces Gut Microbiota Dysbiosis and Increases Systemic Inflammation in Aged Mice
Abstract
:1. Introduction
2. Results
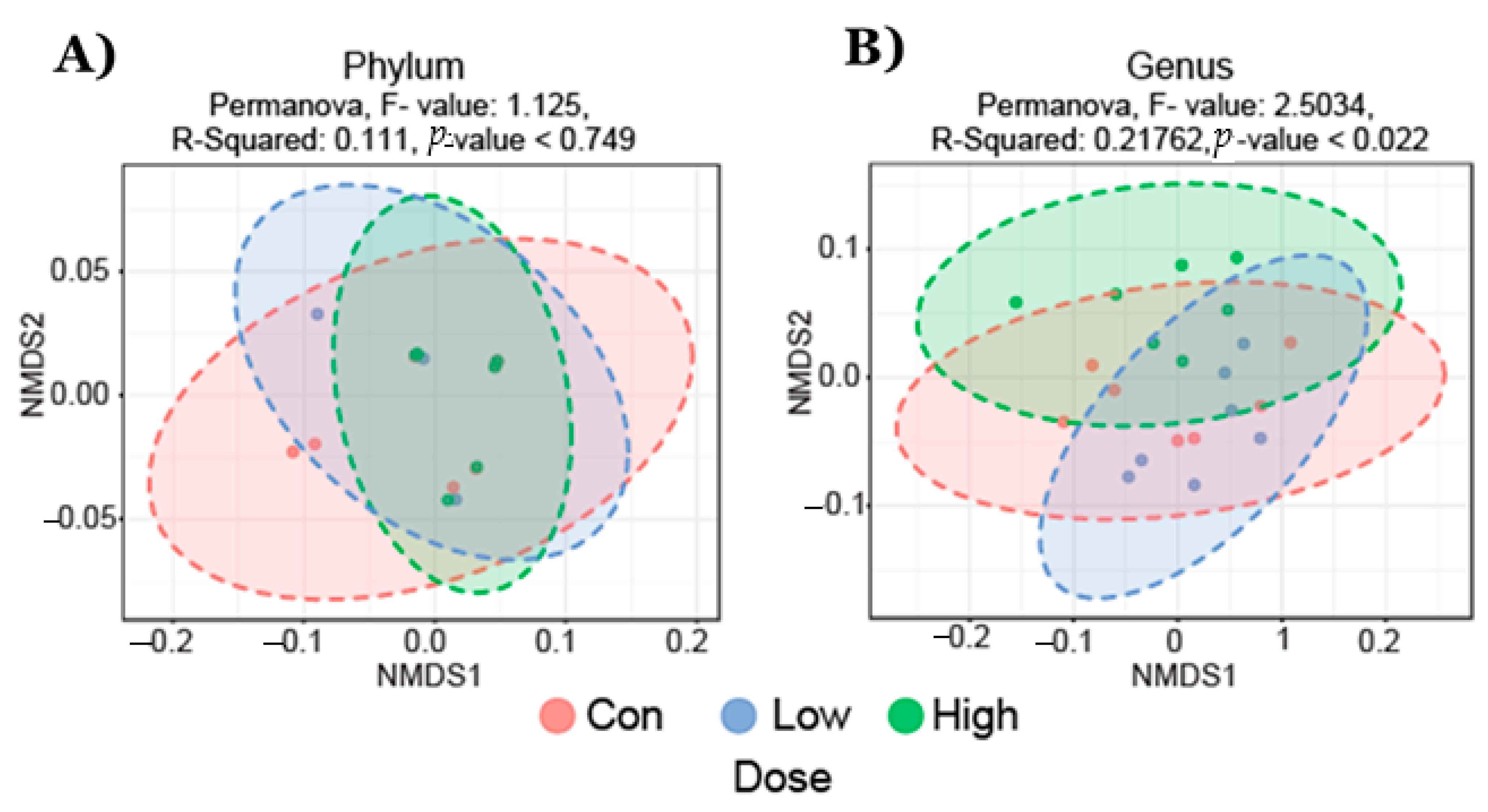
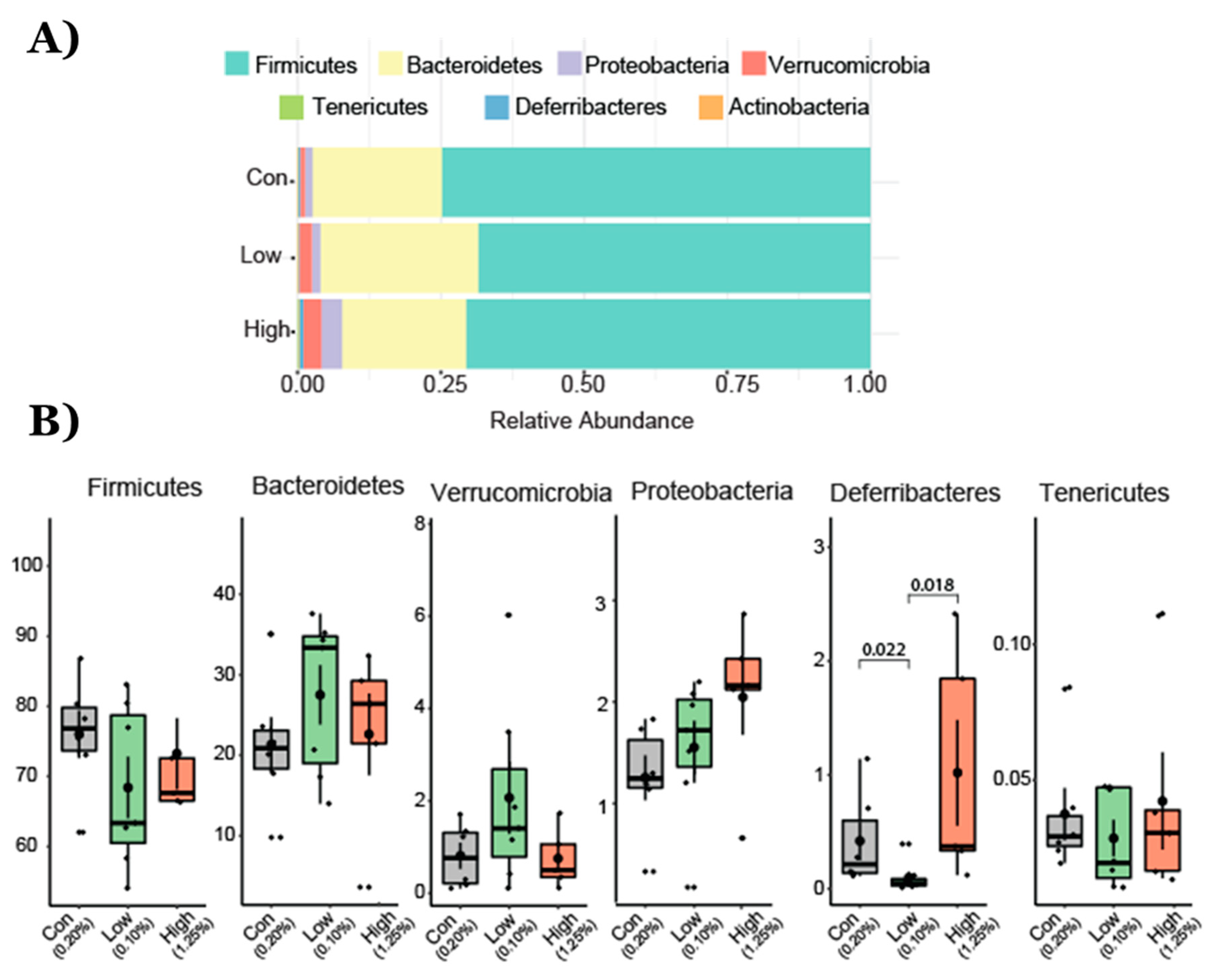
2.1. Gut Microbial Taxonomic Analysis
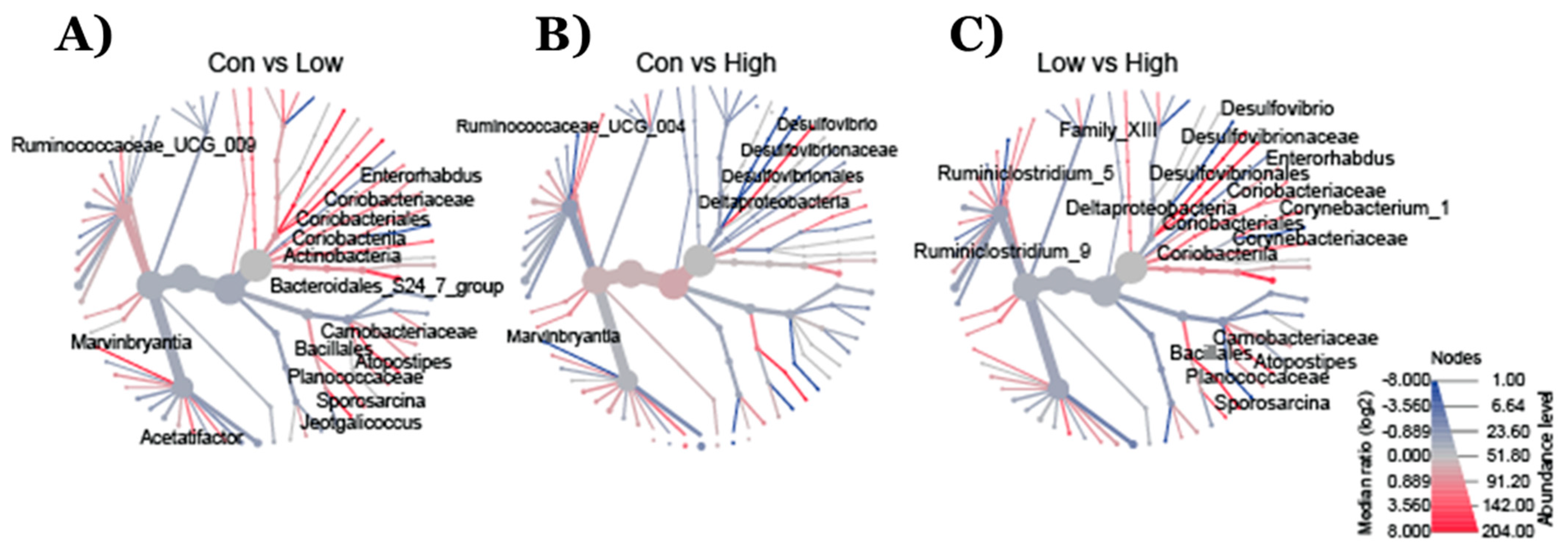
2.2. Effect of TRP Supplementation on Taxonomical Differences
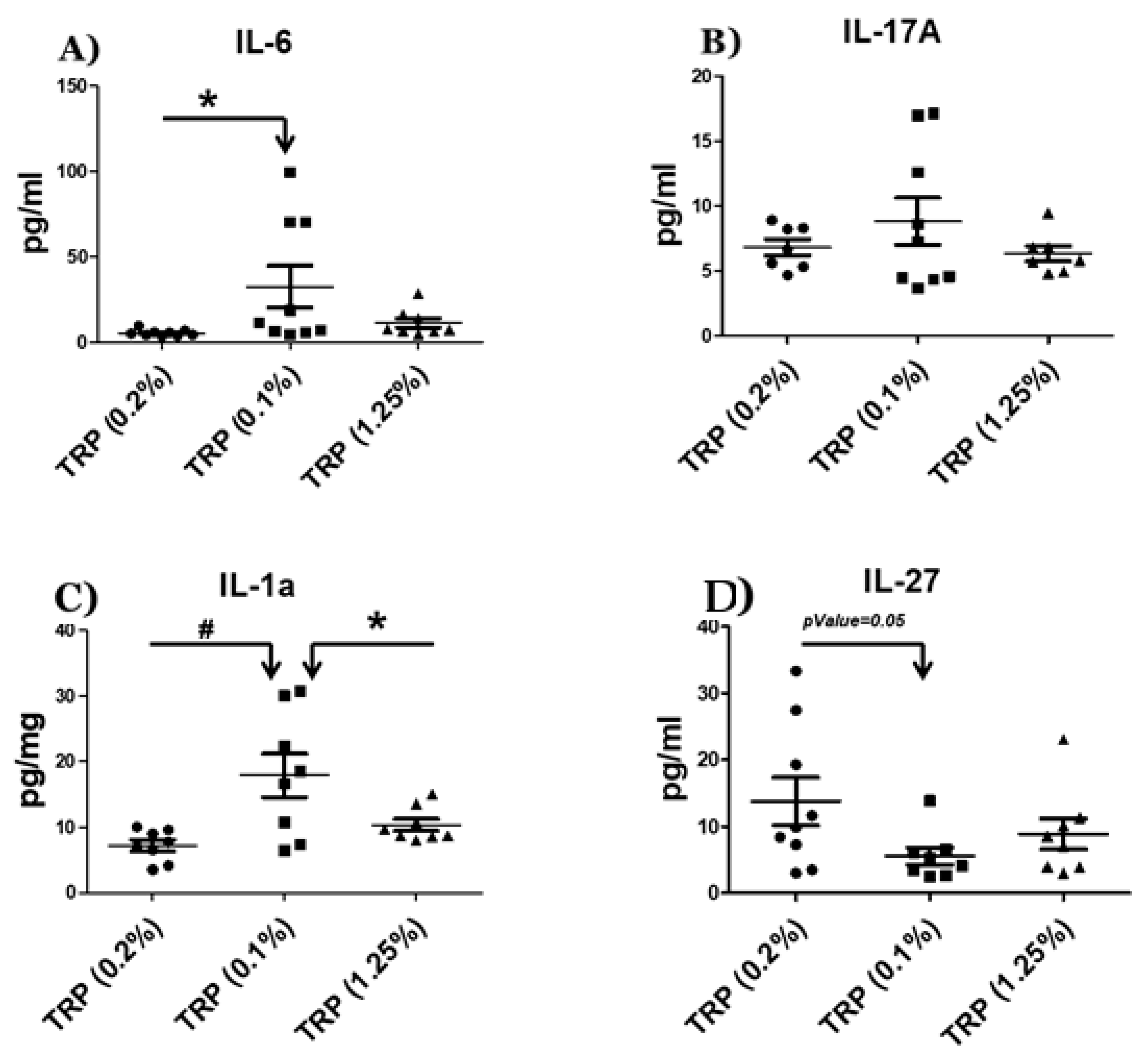
2.3. Serum Cytokine Analysis
3. Discussion
4. Materials and Methods
4.1. Animal Study
4.2. Microbial Community Profiling Using 16S rRNA Amplicon Sequencing
4.3. Bioinformatics Analysis
4.4. Serum Cytokine Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GI | Gastrointestinal |
| AA | Amino acid |
| TRP | Tryptophan |
| IS | Indoxyl sulfate |
| IPA | IPA; Indole-3-propionic acid |
| IDO1 | IDO1; Indoleamine 2,3-dioxygenase |
| AhR | AhR; Aryl hydrocarbon receptor |
References
- Shlisky, J.; Bloom, D.E.; Beaudreault, A.R.; Tucker, K.L.; Keller, H.H.; Freund-Levi, Y.; Fielding, R.A.; Cheng, F.W.; Jensen, G.L.; Wu, D.; et al. Nutritional considerations for healthy aging and reduction in age-related chronic disease. Adv. Nutr. 2017, 8, 17–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakodkar, S.; Mutlu, E.A. Diet as a Therapeutic option for adult inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2017, 46, 745–767. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gol, S.; Pena, R.N.; Rothschild, M.F.; Tor, M.; Estany, J. Dietary tryptophan deficiency and its supplementation compromises inflammatory mechanisms and disease resistance in a teleost fish. Sci. Rep. 2019, 9, 7689. [Google Scholar] [CrossRef]
- Benkelfat, C.; Ellenbogen, M.A.; Dean, P.; Palmour, R.M.; Young, S.N. Mood-lowering effect of tryptophan depletion. Enhanced susceptibility in young men at genetic risk for major affective disorders. Arch. Gen. Psychiatry 1994, 51, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Coppen, A.; Eccleston, E.G.; Peet, M. Total and free tryptophan concentration in the plasma of depressive patients. Lancet 1973, 2, 60–63. [Google Scholar] [CrossRef]
- Henderson, L.M.; Koeppe, O.J.; Zimmerman, H.H. Niacin-tryptophan deficiency resulting from amono acid imbalance in non-casein diets. J. Biol. Chem. 1953, 201, 697–706. [Google Scholar] [CrossRef]
- Hori, M.; Fisher, J.M.; Rabinovitz, M. Tryptophan deficiency in rabbit reticulocytes: Polyribosomes during interrupted growth of hemoglobin chains. Science 1967, 155, 83–84. [Google Scholar] [CrossRef]
- Konopelski, P.; Konop, M.; Gawrys-Kopczynska, M.; Podsadni, P.; Szczepanska, A.; Ufnal, M. Indole-3-propionic acid, a tryptophan-derived bacterial metabolite, reduces weight gain in rats. Nutrients 2019, 11, 591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and Signaling pathways of amino acids in intestinal inflammation. Biomed. Res. Int. 2018, 2018, 9171905. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.; Hong, S.N.; Paik, N.Y.; Kim, T.J.; Kim, E.R.; Chang, D.K.; Kim, Y.H. CD1d modulates colonic inflammation in NOD2-/-mice by altering the intestinal microbial composition comprising acetatifactor muris. J. Crohns Colitis 2019, 13, 1081–1091. [Google Scholar] [CrossRef]
- Ai, D.; Pan, H.; Li, X.; Gao, Y.; Liu, G.; Xia, L.C. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front. Microbiol. 2019, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.Y.; Zolnik, C.P.; Wang, Z.; Qiu, Y.; Usyk, M.; Wang, T.; Kizer, J.R.; Landay, A.L.; Kurland, I.J.; Anastos, K.; et al. Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine 2018, 37, 392–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Adesso, S.; Ruocco, M.; Rapa, S.F.; Dal Piaz, F.; Di Iorio, B.R.; Popolo, A.; Nishijima, F.; Pinto, A.; Marzocco, S. Effect of indoxyl sulfate on the repair and intactness of intestinal epithelial cells: Role of reactive oxygen species’ release. Int. J. Mol. Sci. 2019, 20, 2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondot, S.; Kang, S.; Furet, J.P.; Aguirre de Cárcer, D.; McSweeney, C.; Morrison, M.; Marteau, P.; Dore, J.; Leclerc, M. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm. Bowel Dis. 2011, 17, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diegelmann, J.; Olszak, T.; Goke, B.; Blumberg, R.S.; Brand, S. A novel role for interleukin-27 (IL-27) as mediator of intestinal epithelial barrier protection mediated via differential signal transducer and activator of transcription (STAT) protein signaling and induction of antibacterial and anti-inflammatory proteins. J. Biol. Chem. 2012, 287, 286–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghi, M.; Puccetti, M.; Pariano, M.; Renga, G.; Stincardini, C.; Ricci, M.; Giovagnoli, S.; Costantini, C.; Romani, L. Tryptophan as a central hub for host/microbial symbiosis. Int. J. Tryptophan Res. 2020, 13, 1178646920919755. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and tryptophan metabolism: Endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 2015, 43, 1522–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; He, T.; Johnston, L.J.; Ma, X. Host-microbiome interactions: The aryl hydrocarbon receptor as a critical node in tryptophan metabolites to brain signaling. Gut Microbes 2020, 11, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; Gensollen, T.; Gandhi, A.; Oh, S.F.; Neves, J.F.; Collin, F.; Lavin, R.; Serra, C.; Glickman, J.; de Silva, P.S.A.; et al. dietary and microbial oxazoles induce intestinal inflammation by modulating aryl hydrocarbon receptor responses. Cell 2018, 173, 1123–1134.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chyan, Y.J.; Poeggeler, B.; Omar, R.A.; Chain, D.G.; Frangione, B.; Ghiso, J.; Pappolla, M.A. Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J. Biol. Chem. 1999, 274, 21937–21942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendheim, P.E.; Poeggeler, B.; Neria, E.; Ziv, V.; Pappolla, M.A.; Chain, D.G. Development of indole-3-propionic acid (OXIGON) for Alzheimer’s disease. J. Mol. Neurosci. 2002, 19, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.H.; Xin, F.Z.; Xue, Y.; Hu, Z.; Han, Y.; Ma, F.; Zhou, D.; Liu, X.L.; Cui, A.; Liu, Z.; et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, H.W.; Cui, M.; Li, Y.; Dong, J.L.; Zhang, S.Q.; Zhu, C.C.; Jiang, M.; Zhu, T.; Wang, B.; Wang, H.C.; et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome 2020, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Negatu, D.A.; Liu, J.J.J.; Zimmerman, M.; Kaya, F.; Dartois, V.; Aldrich, C.C.; Gengenbacher, M.; Dick, T. Whole-cell screen of fragment library identifies gut microbiota metabolite indole propionic acid as antitubercular. Antimicrob. Agents Chemother. 2018, 62, e01571-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorgdrager, F.J.H.; Naudé, P.J.W.; Kema, I.P.; Nollen, E.A.; Deyn, P.P. Tryptophan metabolism in inflammaging: From biomarker to therapeutic target. Front. Immunol. 2019, 10, 2565. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and extreme longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.A.; Patterson, A.D.; Perdew, G.H. Aryl hydrocarbon receptor ligands in cancer: Friend and foe. Nat. Rev. Cancer 2014, 14, 801–814. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refaey, M.E.; McGee-Lawrence, M.E.; Fulzele, S.; Kennedy, E.J.; Bollag, W.B.; Elsalanty, M.; Zhong, Q.; Ding, K.H.; Bendzunas, N.G.; Shi, X.M.; et al. Kynurenine, a Tryptophan metabolite that accumulates with age, induces bone loss. J. Bone Miner. Res. 2017, 32, 2182–2193. [Google Scholar] [CrossRef] [PubMed]
- Sanz, H.; Aponte, J.J.; Harezlak, J.; Dong, Y.; Ayestaran, A.; Nhabomba, A.; Mpina, M.; Maurin, O.R.; Díez-Padrisa, N.; Aguilar, R.; et al. drLumi: An open-source package to manage data, calibrate, and conduct quality control of multiplex bead-based immunoassays data analysis. PLoS ONE 2017, 12, e0187901. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yusufu, I.; Ding, K.; Smith, K.; Wankhade, U.D.; Sahay, B.; Patterson, G.T.; Pacholczyk, R.; Adusumilli, S.; Hamrick, M.W.; Hill, W.D.; et al. A Tryptophan-Deficient Diet Induces Gut Microbiota Dysbiosis and Increases Systemic Inflammation in Aged Mice. Int. J. Mol. Sci. 2021, 22, 5005. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095005
Yusufu I, Ding K, Smith K, Wankhade UD, Sahay B, Patterson GT, Pacholczyk R, Adusumilli S, Hamrick MW, Hill WD, et al. A Tryptophan-Deficient Diet Induces Gut Microbiota Dysbiosis and Increases Systemic Inflammation in Aged Mice. International Journal of Molecular Sciences. 2021; 22(9):5005. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095005
Chicago/Turabian StyleYusufu, Ibrahim, Kehong Ding, Kathryn Smith, Umesh D. Wankhade, Bikash Sahay, G. Taylor Patterson, Rafal Pacholczyk, Satish Adusumilli, Mark W. Hamrick, William D. Hill, and et al. 2021. "A Tryptophan-Deficient Diet Induces Gut Microbiota Dysbiosis and Increases Systemic Inflammation in Aged Mice" International Journal of Molecular Sciences 22, no. 9: 5005. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095005






