Estradiol Regulates mRNA Levels of Estrogen Receptor Beta 4 and Beta 5 Isoforms and Modulates Human Granulosa Cell Apoptosis
Abstract
:1. Introduction
2. Results
2.1. Positive Correlations between Pairs of ERβ Isoform mRNA Levels in hGCs
2.2. E2 Promotes Estrogen Receptor Transactivations in hGCs
2.3. E2 Selectively Increased mRNA Levels of ERβ4 and ERβ5 Isoforms in hGCs
2.4. Absence of Influence of ERβ2, ERβ4, or ERβ5 on E2-Induced ERβ1 Transactivation in HGrC1-Transfected Cells
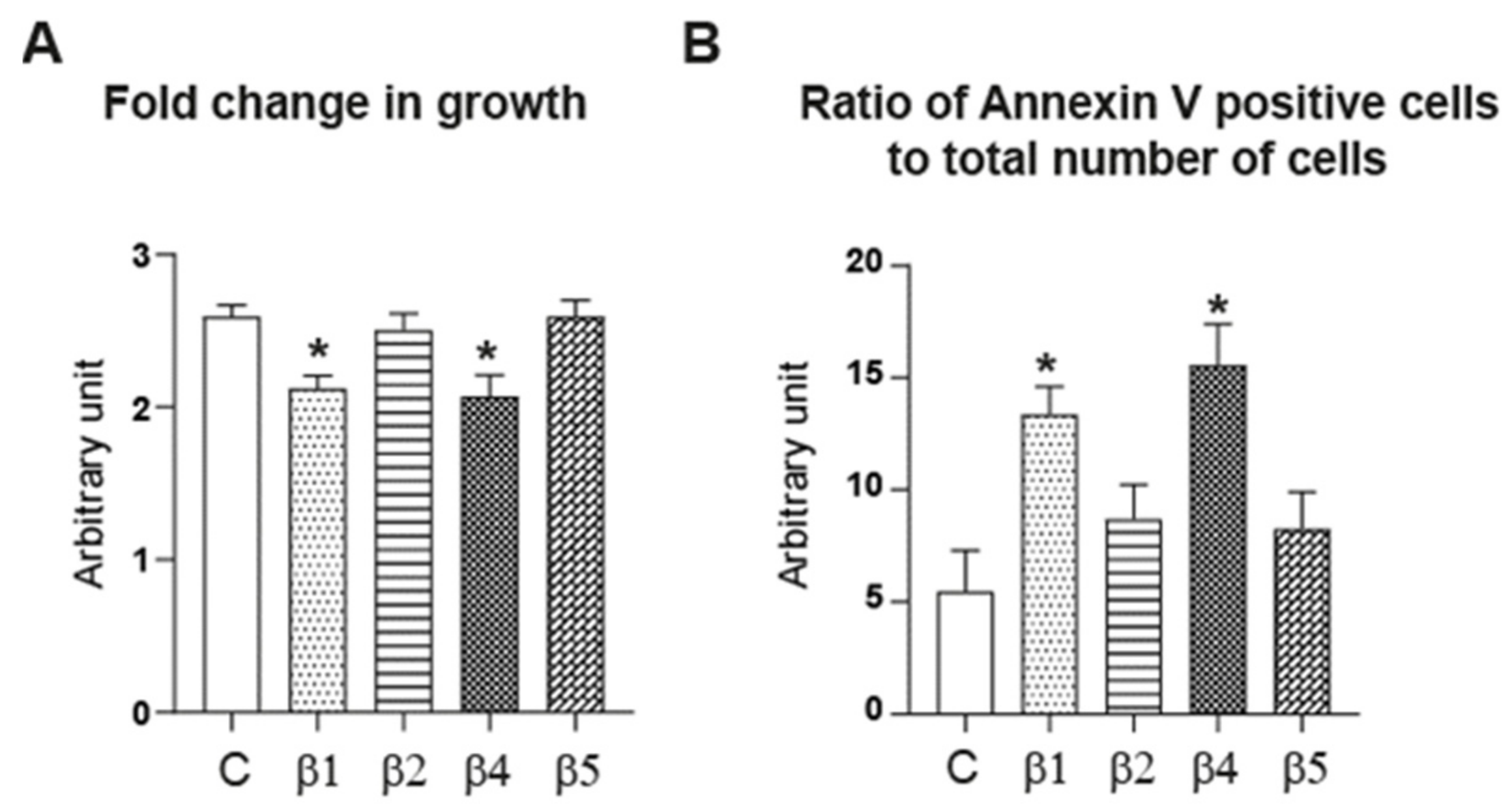
2.5. ERβ1, ERβ2, ERβ4, and ERβ5 Differentially Affect HGrC1 Apoptosis
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Human Granulosa-Lutein Cells Isolation, Culture, and Treatment
4.3. HGrC1 Cells Culture
4.4. Construction of FLAG-Tagged Fusion Protein Expression Vectors
4.5. Transactivation Assays of HGrC1 Cells and Human Primary Granulosa Cells
4.5.1. HGrC1 Cells
4.5.2. hGCs
4.6. Nucleofection of HGrC1 Cells
4.7. Cell Viability and Apoptosis Assays
4.7.1. Cell Viability MTT Assay
4.7.2. Early Apoptosis Assay
4.8. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
| Target Gene (Protein) Accession No. | Forward Primer (Tm) Reverse Primer (Tm) | Product Size, Amplification Efficiency |
|---|---|---|
| ESR1 (ERα) NM_001122740 | 5’-CCA CCA ACC AGT GCA CCA TT-3’ (59.4 °C) 5’-GGT CTT TTC GTA TCC CAC CTT TC-3’ (60.6 °C) | 116 bp, 97.2% |
| ESR2 v1 (ERβ1) NM_001437 | 5’-GTC AGG CAT GCG AGT AAC AA-3’ (57.3 °C) 5’-GGG AGC CCT CTT TGC TTT TA-3’ (57.3 °C) | 192 bp, 90.9% |
| ESR2 v2 (ERβ2) NM_001040275 | 5’-TCT CCT CCC AGC AGC AAT CC-3’ (61.4 °C) 5’-GGT CAC TGC TCC ATC GTT GC-3’ (61.4 °C) | 162 bp, 96% |
| ESR2 v4 (ERβ4) NM_001214902 | 5’- GTG ACC GAT GCT TTG GTT TG-3’ (57.3 °C) 5’-ATC TTT CAT TGC CCA CAT GC-3’ (55.3 °C) | 210 bp, 96% |
| ESR2 v5 (ERβ5) DQ838583.1 | 5’-GAT GCT TTG GTT TGG GTG AT-3’ (55.3 °C) 5’-CCT CCG TGG AGC ACA TAA TC-3’ (59.4 °C) | 177 bp, 95% |
| GAPDH (GAPDH) NM_002046.7 | 5’-TCC CTG AGC TGA ACG GGA AG-3’ (61.4 °C) 5’-GGA GGA GTG GGT GTC GCT GT-3’ (63.5 °C) | 227 bp, 98.5% |
4.9. Immunofluorescence of Transfected HGrC1 Cells
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manabe, N.; Goto, Y.; Matsuda-Minehata, F.; Inoue, N.; Maeda, A.; Sakamaki, K.; Miyano, T. Regulation mechanism of selective atresia in porcine follicles: Regulation of granulosa cell apoptosis during atresia. J. Reprod. Dev. 2004, 50, 493–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNatty, K.P.; Moore Smith, D.; Osathanondh, R.; Ryan, K.J. The human antral follicle: Functional correlates of growth and atresia. Ann. Biol. Anim. Bioch. Biophys. 1979, 19, 1547–1558. [Google Scholar] [CrossRef]
- Fortune, J.E.; Rivera, G.M.; Yang, M.Y. Follicular development: The role of the follicular microenvironment in selection of the dominant follicle. Anim. Reprod. Sci. 2004, 82–83, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.S.; Sui, H.S.; Han, Z.B.; Li, W.; Luo, M.J.; Tan, J.H. Apoptosis in granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004, 14, 341–346. [Google Scholar] [CrossRef] [Green Version]
- Hutz, R.J.; Dierschke, D.J.; Wolf, R.C. Estradiol-induced follicular atresia in rhesus monkeys is not prevented by exogenous gonadotropins. Am. J. Primatol. 1991, 23, 247–255. [Google Scholar] [CrossRef]
- Hutz, R.J.; Dierschke, D.J.; Wolf, R.C. Role of estradiol in regulating ovarian follicular atresia in rhesus monkeys: A review. J. Med. Primatol. 1990, 19, 553–571. [Google Scholar] [CrossRef]
- Billig, H.; Furuta, I.; Hsueh, A.J. Estrogens inhibit and androgens enhance ovarian granulosa cell apoptosis. Endocrinology 1993, 133, 2204–2212. [Google Scholar] [CrossRef]
- Hegele-Hartung, C.; Siebel, P.; Peters, O.; Kosemund, D.; Muller, G.; Hillisch, A.; Walter, A.; Kraetzschmar, J.; Fritzemeier, K.H. Impact of isotype-selective estrogen receptor agonists on ovarian function. Proc. Natl. Acad. Sci. USA 2004, 101, 5129–5134. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.A.; Saunders, P.T. Estrogen dependent signaling in reproductive tissues—A role for estrogen receptors and estrogen related receptors. Mol. Cell. Endocrinol. 2012, 348, 361–372. [Google Scholar] [CrossRef]
- Mueller, S.O.; Katzenellenbogen, J.A.; Korach, K.S. Endogenous estrogen receptor beta is transcriptionally active in primary ovarian cells from estrogen receptor knockout mice. Steroids 2004, 69, 681–686. [Google Scholar] [CrossRef]
- Donoghue, L.J.; Neufeld, T.I.; Li, Y.; Arao, Y.; Coons, L.A.; Korach, K.S. Differential Activation of a Mouse Estrogen Receptor beta Isoform (mERbeta2) with Endocrine-Disrupting Chemicals (EDCs). Environ. Health Perspect 2017, 125, 634–642. [Google Scholar] [CrossRef] [Green Version]
- Scobie, G.A.; Macpherson, S.; Millar, M.R.; Groome, N.P.; Romana, P.G.; Saunders, P.T. Human oestrogen receptors: Differential expression of ER alpha and beta and the identification of ER beta variants. Steroids 2002, 67, 985–992. [Google Scholar] [CrossRef]
- Poola, I. Molecular assays to profile 10 estrogen receptor beta isoform mRNA copy numbers in ovary, breast, uterus, and bone tissues. Endocrine 2003, 22, 101–112. [Google Scholar] [CrossRef]
- Moore, J.T.; McKee, D.D.; Slentz-Kesler, K.; Moore, L.B.; Jones, S.A.; Horne, E.L.; Su, J.L.; Kliewer, S.A.; Lehmann, J.M.; Willson, T.M. Cloning and characterization of human estrogen receptor beta isoforms. Biochem. Biophys. Res. Commun. 1998, 247, 75–78. [Google Scholar] [CrossRef]
- Leung, Y.K.; Mak, P.; Hassan, S.; Ho, S.M. Estrogen receptor (ER)-beta isoforms: A key to understanding ER-beta signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 13162–13167. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Gustafsson, J.A. The different roles of ER subtypes in cancer biology and therapy. Nat. Rev. Cancer 2011, 11, 597–608. [Google Scholar] [CrossRef]
- Collins, F.; Itani, N.; Esnal-Zufiaurre, A.; Gibson, D.A.; Fitzgerald, C.; Saunders, P.T.K. The ERbeta5 splice variant increases oestrogen responsiveness of ERalphapos Ishikawa cells. Endocr. Relat. Cancer 2020, 27, 55–66. [Google Scholar] [CrossRef]
- Peng, B.; Lu, B.; Leygue, E.; Murphy, L.C. Putative functional characteristics of human estrogen receptor-beta isoforms. J. Mol. Endocrinol. 2003, 30, 13–29. [Google Scholar] [CrossRef] [Green Version]
- Poola, I.; Abraham, J.; Baldwin, K.; Saunders, A.; Bhatnagar, R. Estrogen receptors beta4 and beta5 are full length functionally distinct ERbeta isoforms: Cloning from human ovary and functional characterization. Endocrine 2005, 27, 227–238. [Google Scholar] [CrossRef]
- Zhao, C.; Matthews, J.; Tujague, M.; Wan, J.; Strom, A.; Toresson, G.; Lam, E.W.; Cheng, G.; Gustafsson, J.A.; Dahlman-Wright, K. Estrogen receptor beta2 negatively regulates the transactivation of estrogen receptor alpha in human breast cancer cells. Cancer Res. 2007, 67, 3955–3962. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, J.; Iwasaki, S.; Yanaihara, A.; Negishi, M.; Tahara, R.; Okai, T. Correlations between steroids concentration in follicular fluid, pronuclear morphology and embryo qualities in in vitro fertilization. Reprod. Med. Biol. 2003, 2, 171–176. [Google Scholar] [CrossRef]
- Yding Andersen, C.; Rosendahl, M.; Byskov, A.G. Concentration of anti-Mullerian hormone and inhibin-B in relation to steroids and age in follicular fluid from small antral human follicles. J. Clin. Endocrinol. Metab. 2008, 93, 2344–2349. [Google Scholar] [CrossRef] [Green Version]
- Bayasula Iwase, A.; Kiyono, T.; Takikawa, S.; Goto, M.; Nakamura, T.; Nagatomo, Y.; Nakahara, T.; Kotani, T.; Kobayashi, H.; Kondo, M.; et al. Establishment of a human nonluteinized granulosa cell line that transitions from the gonadotropin-independent to the gonadotropin-dependent status. Endocrinology 2012, 153, 2851–2860. [Google Scholar] [CrossRef]
- Hager, G.L.; Lim, C.S.; Elbi, C.; Baumann, C.T. Trafficking of nuclear receptors in living cells. J. Steroid Biochem. Mol. Biol. 2000, 74, 249–254. [Google Scholar] [CrossRef]
- Treeck, O.; Pfeiler, G.; Mitter, D.; Lattrich, C.; Piendl, G.; Ortmann, O. Estrogen receptor β1 exerts antitumoral effects on SK-OV-3 ovarian cancer cells. J. Endocrinol. 2007, 193, 421–433. [Google Scholar] [CrossRef]
- Sikora, M.J.; Johnson, M.D.; Lee, A.V.; Oesterreich, S. Endocrine Response Phenotypes Are Altered by Charcoal-Stripped Serum Variability. Endocrinology 2016, 157, 3760–3766. [Google Scholar] [CrossRef] [Green Version]
- Ko, C.; Meidan, R.; Bridges, P.J. Why two endothelins and two receptors for ovulation and luteal regulation? Life Sci. 2012, 91, 501–506. [Google Scholar] [CrossRef] [Green Version]
- Bridges, P.J.; Cho, J.; Ko, C. Endothelins in regulating ovarian and oviductal function. Front. Biosci. 2011, 3, 145–155. [Google Scholar]
- Choi, D.H.; Kim, E.K.; Kim, K.H.; Lee, K.A.; Kang, D.W.; Kim, H.Y.; Bridges, P.; Ko, C. Expression pattern of endothelin system components and localization of smooth muscle cells in the human pre-ovulatory follicle. Hum. Reprod. 2011, 26, 1171–1180. [Google Scholar] [CrossRef] [Green Version]
- Gohar, E.Y.; Yusuf, C.; Pollock, D.M. Ovarian hormones modulate endothelin A and B receptor expression. Life Sci. 2016, 159, 148–152. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, S.S.; Huang, H.L.; Song, B.J.; Liu, X.; Qi, Z. Enhanced Endothelin A and B Receptor Expression and Receptor-Mediated Vasoconstriction in Rat Mesenteric arteries after Lipopolysaccharide Challenge. Mediat. Inflamm. 2019, 2019, 6248197. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Brannan, R.A.; Hanby, A.M.; Shaaban, A.M.; Verghese, E.T.; Peter, M.B.; Pollock, S.; Satheesha, S.; Szynkiewicz, M.; Speirs, V.; et al. Differential regulation of oestrogen receptor beta isoforms by 5’ untranslated regions in cancer. J. Cell. Mol. Med. 2010, 14, 2172–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, L.; Coleman, L.J.; Cummings, M.; Satheesha, S.; Shaw, S.O.; Speirs, V.; Hughes, T.A. Expression of oestrogen receptor beta isoforms is regulated by transcriptional and post-transcriptional mechanisms. Biochem. J. 2010, 429, 283–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, S.; Shoda, T.; Kato, J.; Hoshi, K. The multiple untranslated first exons system of the human estrogen receptor beta (ER beta) gene. J. Steroid Biochem. Mol. Biol. 2001, 78, 33–40. [Google Scholar] [CrossRef]
- Lee, M.T.; Ouyang, B.; Ho, S.M.; Leung, Y.K. Differential expression of estrogen receptor beta isoforms in prostate cancer through interplay between transcriptional and translational regulation. Mol. Cell. Endocrinol. 2013, 376, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Adams, B.D.; Furneaux, H.; White, B.A. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen recep-tor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007, 21, 1132–1147. [Google Scholar] [CrossRef]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef] [Green Version]
- Al-Nakhle, H.; Burns, P.A.; Cummings, M.; Hanby, A.M.; Hughes, T.A.; Satheesha, S.; Shaaban, A.M.; Smith, L.; Speirs, V. Estrogen receptor β1 expression is regulated by miR-92 in breast cancer. Cancer Res. 2010, 70, 4778–4784. [Google Scholar] [CrossRef] [Green Version]
- Weihua, Z.; Ekman, J.; Almkvist, A.; Saji, S.; Wang, L.; Warner, M.; Gustafsson, J.A. Involvement of androgen receptor in 17beta-estradiol-induced cell proliferation in rat uterus. Biol. Reprod. 2002, 67, 616–623. [Google Scholar] [CrossRef] [Green Version]
- Gava, N.; Clarke, C.L.; Byth, K.; Arnett-Mansfield, R.L.; de Fazio, A. Expression of progesterone receptors A and B in the mouse ovary during the estrous cycle. Endocrinology 2004, 145, 3487–3494. [Google Scholar] [CrossRef]
- Zhang, J.; Lazar, M.A. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 2000, 62, 439–466. [Google Scholar] [CrossRef]
- Glass, C.K.; Holloway, J.M.; Devary, O.V.; Rosenfeld, M.G. The thyroid hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid hormone and estrogen response elements. Cell 1988, 54, 313–323. [Google Scholar] [CrossRef]
- Dey, P.; Jonsson, P.; Hartman, J.; Williams, C.; Strom, A.; Gustafsson, J.A. Estrogen receptors beta1 and beta2 have opposing roles in regulating proliferation and bone metastasis genes in the prostate cancer cell line PC3. Mol. Endocrinol. 2012, 26, 1991–2003. [Google Scholar] [CrossRef]
- Langdon, S.P.; Herrington, C.S.; Hollis, R.L.; Gourley, C. Estrogen Signaling and Its Potential as a Target for Therapy in Ovarian Cancer. Cancers 2020, 12, 1647. [Google Scholar] [CrossRef]
- Liu, J.; Sareddy, G.R.; Zhou, M.; Viswanadhapalli, S.; Li, X.; Lai, Z.; Tekmal, R.R.; Brenner, A.; Vadlamudi, R.K. Differential Effects of Estrogen Receptor beta Isoforms on Glioblastoma Progression. Cancer Res. 2018, 78, 3176–3189. [Google Scholar] [CrossRef] [Green Version]
- Ajmal, N.; Khan, S.Z.; Shaikh, R. Polycystic ovary syndrome (PCOS) and genetic predisposition: A review article. Eur. J. Obs. Gynecol. Reprod. Biol. X 2019, 3, 100060. [Google Scholar] [CrossRef]
- Das, M.; Djahanbakhch, O.; Hacihanefioglu, B.; Saridogan, E.; Ikram, M.; Ghali, L.; Raveendran, M.; Storey, A. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2008, 93, 881–887. [Google Scholar] [CrossRef] [Green Version]
- Owens, L.A.; Kristensen, S.G.; Lerner, A.; Christopoulos, G.; Lavery, S.; Hanyaloglu, A.C.; Hardy, K.; Yding Andersen, C.; Franks, S. Gene Expression in Granulosa Cells from Small Antral Follicles From Women With or Without Polycystic Ovaries. J. Clin. Endocrinol. Metab. 2019, 104, 6182–6192. [Google Scholar] [CrossRef] [Green Version]
- Artimani, T.; Saidijam, M.; Aflatoonian, R.; Amiri, I.; Ashrafi, M.; Shabab, N.; Mohammadpour, N.; Mehdizadeh, M. Estrogen and progesterone receptor subtype expression in granulosa cells from women with polycystic ovary syndrome. Gynecol. Endocrinol. 2015, 31, 379–383. [Google Scholar] [CrossRef]
- Pierre, A.; Taieb, J.; Giton, F.; Grynberg, M.; Touleimat, S.; El Hachem, H.; Fanchin, R.; Monniaux, D.; Cohen-Tannoudji, J.; di Clemente, N.; et al. Dysregulation of the Anti-Mullerian Hormone System by Steroids in Women with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2017, 102, 3970–3978. [Google Scholar] [CrossRef] [Green Version]
- Jakimiuk, A.J.; Weitsman, S.R.; Yen, H.W.; Bogusiewicz, M.; Magoffin, D.A. Estrogen receptor alpha and beta expression in theca and granulosa cells from women with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2002, 87, 5532–5538. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierre, A.; Mayeur, A.; Marie, C.; Cluzet, V.; Chauvin, J.; Frydman, N.; Grynberg, M.; Cohen-Tannoudji, J.; Guigon, C.J.; Chauvin, S. Estradiol Regulates mRNA Levels of Estrogen Receptor Beta 4 and Beta 5 Isoforms and Modulates Human Granulosa Cell Apoptosis. Int. J. Mol. Sci. 2021, 22, 5046. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095046
Pierre A, Mayeur A, Marie C, Cluzet V, Chauvin J, Frydman N, Grynberg M, Cohen-Tannoudji J, Guigon CJ, Chauvin S. Estradiol Regulates mRNA Levels of Estrogen Receptor Beta 4 and Beta 5 Isoforms and Modulates Human Granulosa Cell Apoptosis. International Journal of Molecular Sciences. 2021; 22(9):5046. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095046
Chicago/Turabian StylePierre, Alice, Anne Mayeur, Clémentine Marie, Victoria Cluzet, Jonathan Chauvin, Nelly Frydman, Michael Grynberg, Joelle Cohen-Tannoudji, Céline J. Guigon, and Stéphanie Chauvin. 2021. "Estradiol Regulates mRNA Levels of Estrogen Receptor Beta 4 and Beta 5 Isoforms and Modulates Human Granulosa Cell Apoptosis" International Journal of Molecular Sciences 22, no. 9: 5046. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22095046






