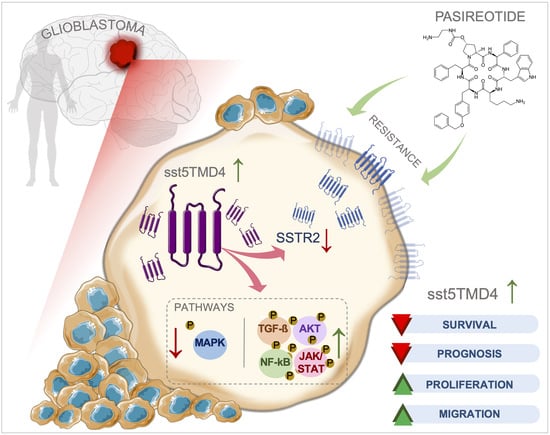
Somatostatin Receptor Splicing Variant sst5TMD4 Overexpression in Glioblastoma Is Associated with Poor Survival, Increased Aggressiveness Features, and Somatostatin Analogs Resistance
Abstract
:1. Introduction
2. Results
2.1. sst5TMD4 Levels Are Significantly Overexpressed in GBM Samples and Associated with Poor Prognosis and Survival Rate in Patients with GBM
2.2. Overexpression of sst5TMD4 Increases Aggressiveness Parameters in GBM Cells
2.3. Overexpression of sst5TMD4 Modulated the Phosphorylation Levels of Different Proteins Associated with Key Oncogenic Pathways in GBM Cells
2.4. Silencing of sst5TMD4 Reduces Aggressiveness Parameters in GBM Cells
2.5. Overexpression of sst5TMD4 Alters the Basal Expression of SSTR2 in GBM Cells and Sensitized the Response of GBM Cells to Pasireotide Treatment
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Patients and Samples
4.3. Immunohistochemical Analysis
4.4. Cell Cultures
4.5. Modulation of the Levels of sst5TMD4 Expression (Overexpression and Silencing) in GBM Cells
4.6. Measurements of Cell Proliferation
4.7. Measurement of Cell Migration Capacity
4.8. RNA Extraction, Retrotranscription, and Gene Expression Measurement by qPCR
4.9. Human Phosphorylation Pathway Profiling Array
4.10. Bioinformatic and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro-Oncology 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Ostrom, Q.T.; Kruchko, C.; Patil, N.; Tihan, T.; Cioffi, G.; Fuchs, H.E.; Waite, K.A.; Jemal, A.; Siegel, R.L.; et al. Brain and other central nervous system tumor statistics, 2021. CA A Cancer J. Clin. 2021, 71, 381–406. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Janjua, T.I.; Rewatkar, P.; Ahmed-Cox, A.; Saeed, I.; Mansfeld, F.M.; Kulshreshtha, R.; Kumeria, T.; Ziegler, D.S.; Kavallaris, M.; Mazzieri, R.; et al. Frontiers in the treatment of glioblastoma: Past, present and emerging. Adv. Drug Deliv. Rev. 2021, 171, 108–138. [Google Scholar] [CrossRef]
- Lowe, S.; Bhat, K.P.; Olar, A. Current clinical management of patients with glioblastoma. Cancer Rep. 2019, 2, e1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Martín, B.; Medina, M. Advances in the Knowledge of the Molecular Biology of Glioblastoma and Its Impact in Patient Diagnosis, Stratification, and Treatment. Adv. Sci. 2020, 7, 1902971. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Borrego, M.C.; Fuentes-Fayos, A.C.; Moreno, V.; Rivero-Cortés, E.; Dios, E.; Moreno, M.; Madrazo-Atutxa, A.; Remón-Ruiz, P.; Solivera, J.; Wildemberg, L.E.; et al. Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features. Cancers 2019, 11, 1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentes-Fayos, A.C.; Vázquez-Borrego, M.C.; Jiménez-Vacas, J.M.; Bejarano, L.; Pedraza-Arévalo, S.; L.-López, F.; Blanco-Acevedo, C.; Sánchez-Sánchez, R.; Reyes, O.; Ventura, S.; et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness: Oncogenic role of SRSF3. Brain 2020, 143, 3273–3293. [Google Scholar] [CrossRef]
- Jiménez-Vacas, J.M.; Herrero-Aguayo, V.; Hidalgo, A.J.M.; Gómez-Gómez, E.; Fuentes-Fayos, A.C.; León-González, A.J.; Sáez-Martínez, P.; Alors-Pérez, E.; Pedraza-Arévalo, S.; González-Serrano, T.; et al. Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer. eBioMedicine 2020, 51, 102547. [Google Scholar] [CrossRef] [Green Version]
- Reyes, O.; Pérez, E.; Luque, R.M.; Castaño, J.; Ventura, S. A supervised machine learning-based methodology for analyzing dysregulation in splicing machinery: An application in cancer diagnosis. Artif. Intell. Med. 2020, 108, 101950. [Google Scholar] [CrossRef]
- Günther, T.; Tulipano, G.; Dournaud, P.; Bousquet, C.; Csaba, Z.; Kreienkamp, H.-J.; Lupp, A.; Korbonits, M.; Castaño, J.P.; Wester, H.-J.; et al. International Union of Basic and Clinical Pharmacology. CV. Somatostatin Receptors: Structure, Function, Ligands, and New Nomenclature. Pharmacol. Rev. 2018, 70, 763–835. [Google Scholar] [CrossRef] [Green Version]
- Durán-Prado, M.; Gahete, M.D.; Martínez-Fuentes, A.J.; Luque, R.M.; Quintero, A.; Webb, S.M.; Benito-López, P.; Leal, A.; Schulz, S.; Gracia-Navarro, F.; et al. Identification and Characterization of Two Novel Truncated but Functional Isoforms of the Somatostatin Receptor Subtype 5 Differentially Present in Pituitary Tumors. J. Clin. Endocrinol. Metab. 2009, 94, 2634–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Córdoba-Chacón, J.; Gahete, M.D.; Durán-Prado, M.; Luque, R.M.; Castaño, J.P. Truncated somatostatin receptors as new players in somatostatin-cortistatin pathophysiology. Ann. N. Y. Acad. Sci. 2011, 1220, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Durán-Prado, M.; Saveanu, A.; Luque, R.M.; Gahete, M.D.; Gracia-Navarro, F.; Jaquet, P.; Dufour, H.; Malagon, M.M.; Culler, M.D.; Barlier, A.; et al. A Potential Inhibitory Role for the New Truncated Variant of Somatostatin Receptor 5, sst5TMD4, in Pituitary Adenomas Poorly Responsive to Somatostatin Analogs. J. Clin. Endocrinol. Metab. 2010, 95, 2497–2502. [Google Scholar] [CrossRef] [Green Version]
- Duran-Prado, M.; Gahete, M.D.; Hergueta-Redondo, M.; Martínez-Fuentes, A.J.; Cordoba-Chacon, J.; Palacios, J.; Gracia-Navarro, F.; Moreno-Bueno, G.; Malagon, M.M.; Luque, R.M.; et al. The new truncated somatostatin receptor variant sst5TMD4 is associated to poor prognosis in breast cancer and increases malignancy in MCF-7 cells. Oncogene 2011, 31, 2049–2061. [Google Scholar] [CrossRef] [Green Version]
- Domingo, M.P.; Luque, R.M.; Reverter, J.L.; López-Sánchez, L.M.; Gahete, M.D.; Culler, M.D.; Díaz-Soto, G.; Lomeña, F.; Squarcia, M.; Mate, J.L.; et al. The Truncated Isoform of Somatostatin Receptor5 (sst5TMD4) Is Associated with Poorly Differentiated Thyroid Cancer. PLoS ONE 2014, 9, e85527. [Google Scholar] [CrossRef]
- Luque, R.M.; Ibáñez-Costa, A.; Neto, L.V.; Taboada, G.F.; Hormaechea-Agulla, D.; Kasuki, L.; Venegas-Moreno, E.; Moreno-Carazo, A.; Gálvez, M.; Soto-Moreno, A.; et al. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett. 2015, 359, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molè, D.; Gentilin, E.; Ibanez-Costa, A.; Gagliano, T.; Gahete, M.D.; Tagliati, F.; Rossi, R.; Pelizzo, M.R.; Pansini, G.; Luque, R.M.; et al. The expression of the truncated isoform of somatostatin receptor subtype 5 associates with aggressiveness in medullary thyroid carcinoma cells. Endocrine 2015, 50, 442–452. [Google Scholar] [CrossRef]
- Sampedro-Núñez, M.; Luque, R.M.; Ramos-Levi, A.M.; Gahete, M.D.; Serrano-Somavilla, A.; Villa-Osaba, A.; Adrados, M.; Ibáñez-Costa, A.; Martin-Perez, E.; Culler, M.D.; et al. Presence of sst5TMD4, a truncated splice variant of the somatostatin receptor subtype 5, is associated to features of increased aggressiveness in pancreatic neuroendocrine tumors. Oncotarget 2015, 7, 6593–6608. [Google Scholar] [CrossRef] [Green Version]
- Gahete, M.D.; Rincón-Fernández, D.; Durán-Prado, M.; Hergueta-Redondo, M.; Ibáñez-Costa, A.; Rojo-Sebastián, A.; Gracia-Navarro, F.; Culler, M.D.; Casanovas, O.; Moreno-Bueno, G.; et al. The truncated somatostatin receptor sst5TMD4 stimulates the angiogenic process and is associated to lymphatic metastasis and disease-free survival in breast cancer patients. Oncotarget 2016, 7, 60110–60122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rio-Moreno, M.; Alors-Perez, E.; de Souza, P.B.; Prados-Gonzalez, M.E.; Castaño, J.P.; Luque, R.M.; Gahete, M.D. Peptides derived from the extracellular domain of the somatostatin receptor splicing variant SST5TMD4 increase malignancy in multiple cancer cell types. Transl. Res. 2019, 211, 147–160. [Google Scholar] [CrossRef]
- Hormaechea-Agulla, D.; Jiménez-Vacas, J.M.; Gómez-Gómez, E.; López, F.L.; Carrasco-Valiente, J.; Valero-Rosa, J.; Moreno, M.M.; Sánchez-Sánchez, R.; Ortega-Salas, R.; Gracia-Navarro, F.; et al. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J. 2017, 31, 4682–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marina, D.; Burman, P.; Klose, M.; Casar-Borota, O.; Luque, R.M.; Castaño, J.P.; Feldt-Rasmussen, U. Truncated somatostatin receptor 5 may modulate therapy response to somatostatin analogues—Observations in two patients with acromegaly and severe headache. Growth Horm. IGF Res. 2015, 25, 262–267. [Google Scholar] [CrossRef]
- Rai, U.; Thrimawithana, T.R.; Valery, C.; Young, S. Therapeutic uses of somatostatin and its analogues: Current view and potential applications. Pharmacol. Ther. 2015, 152, 98–110. [Google Scholar] [CrossRef]
- Stueven, A.K.; Kayser, A.; Wetz, C.; Amthauer, H.; Wree, A.; Tacke, F.; Wiedenmann, B.; Roderburg, C.; Jann, H. Somatostatin Analogues in the Treatment of Neuroendocrine Tumors: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 3049. [Google Scholar] [CrossRef] [Green Version]
- Lamszus, K.; Meyerhof, W.; Westphal, M. Somatostatin and somatostatin receptors in the diagnosis and treatment of gliomas. J. Neuro-Oncol. 1997, 35, 353–364. [Google Scholar] [CrossRef]
- Hauser, P.; Hanzely, Z.; Máthé, D.; Szabó, E.; Barna, G.; Sebestyén, A.; Jeney, A.; Schuler, D.; Fekete, G.; Garami, M. EFFECT OF SOMATOSTATIN ANALOGUE OCTREOTIDE IN MEDULLOBLASTOMA IN XENOGRAFT AND CELL CULTURE STUDY. Pediatr. Hematol. Oncol. 2009, 26, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Graillon, T.; Romano, D.; Defilles, C.; Saveanu, A.; Mohamed, A.; Figarella-Branger, D.; Roche, P.-H.; Fuentes, S.; Chinot, O.; Dufour, H.; et al. Octreotide therapy in meningiomas: In vitro study, clinical correlation, and literature review. J. Neurosurg. 2017, 127, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Cavalla, P.; Schiffer, D. Neuroendocrine tumors in the brain. Ann. Oncol. 2001, 12, S131–S134. [Google Scholar] [CrossRef]
- Held-Feindt, J.; Krisch, B.; Forstreuter, F.; Mentlein, R. Somatostatin receptors in gliomas. J. Physiol. 2000, 94, 251–258. [Google Scholar] [CrossRef]
- Grimm, S.A.; Chamberlain, M.C. Anaplastic astrocytoma. CNS Oncol. 2016, 5, 145–157. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. The definition of primary and secondary glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef] [Green Version]
- Diao, W.; Tong, X.; Yang, C.; Zhang, F.; Bao, C.; Chen, H.; Liu, L.; Li, M.; Ye, F.; Fan, Q.; et al. Behaviors of Glioblastoma Cells in in Vitro Microenvironments. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, J.; Srinageshwar, B.; Dunbar, G.L. Current Therapeutic Strategies for Glioblastoma. Brain Sci. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, K.; Lamberts, S.W.J. Somatostatin analogues in acromegaly and gastroenteropancreatic neuroendocrine tumours: Past, present and future. Endocr. Relat. Cancer 2016, 23, R551–R566. [Google Scholar] [CrossRef] [Green Version]
- Fuentes-Fayos, A.C.; García-Martínez, A.; Herrera-Martínez, A.D.; Jiménez-Vacas, J.M.; Vazquez-Borrego, M.C.; Castaño, J.P.; Picó, A.; Gahete, M.D.; Luque, R.M. Molecular determinants of the response to medical treatment of growth hormone secreting pituitary neuroendocrine tumors. Minerva Endocrinol. 2019, 44, 109–128. [Google Scholar] [CrossRef]
- Kiviniemi, A.; Gardberg, M.; Kivinen, K.; Posti, J.P.; Vuorinen, V.; Sipilä, J.; Rahi, M.; Sankinen, M.; Minn, H. Somatostatin receptor 2A in gliomas: Association with oligodendrogliomas and favourable outcome. Oncotarget 2017, 8, 49123–49132. [Google Scholar] [CrossRef] [Green Version]
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef]
- Ladomery, M. Aberrant Alternative Splicing Is Another Hallmark of Cancer. Int. J. Cell Biol. 2013, 2013, 463786. [Google Scholar] [CrossRef]
- Lo, H.-W.; Zhu, H.; Cao, X.; Aldrich, A.; Ali-Osman, F. A Novel Splice Variant ofGLI1That Promotes Glioblastoma Cell Migration and Invasion. Cancer Res. 2009, 69, 6790–6798. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Shamardani, K.; Babikir, H.; Catalan, F.; Nejo, T.; Chang, S.; Phillips, J.J.; Okada, H.; Diaz, A.A. The evolution of alternative splicing in glioblastoma under therapy. Genome Biol. 2021, 22, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Liu, R.; Qu, X.; Yu, H.; Chu, H.; Zhang, Y.; Zhu, W.; Wu, X.; Gao, H.; Tao, B.; et al. α-Ketoglutarate-Activated NF-κB Signaling Promotes Compensatory Glucose Uptake and Brain Tumor Development. Mol. Cell 2019, 76, 148–162.e7. [Google Scholar] [CrossRef]
- Cahill, K.E.; Morshed, R.A.; Yamini, B. Nuclear factor-κB in glioblastoma: Insights into regulators and targeted therapy. Neuro-Oncology 2015, 18, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, J.; Pagano, M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 2002, 13, 41–47. [Google Scholar] [CrossRef]
- Fresno Vara, J.Á.; Casado, E.; De Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Yang, R.; Wang, M.; Zhang, G.; Li, Y.; Wang, L.; Cui, H. POU2F2 regulates glycolytic reprogramming and glioblastoma progression via PDPK1-dependent activation of PI3K/AKT/mTOR pathway. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, K.; Nandeesha, H.; Dorairajan, L.N.; Ganesh, R.N. Over expression of PI3K-AkT reduces apoptosis and increases prostate size in benign prostatic hyperplasia. Aging Male 2018, 23, 440–446. [Google Scholar] [CrossRef]
- Yue, J.; López, J.M. Understanding MAPK Signaling Pathways in Apoptosis. Int. J. Mol. Sci. 2020, 21, 2346. [Google Scholar] [CrossRef] [Green Version]
- Krishna, K.V.; Dubey, S.K.; Singhvi, G.; Gupta, G.; Kesharwani, P. MAPK pathway: Potential role in glioblastoma multiforme. Interdiscip. Neurosurg. 2020, 23, 100901. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.-J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [Green Version]
- Fleseriu, M.; Petersenn, S. New avenues in the medical treatment of Cushing’s disease: Corticotroph tumor targeted therapy. J. Neuro-Oncol. 2013, 114, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mawrin, C.; Schulz, S.; Pauli, S.U.; Treuheit, T.; Diete, S.; Dietzmann, K.; Firsching, R.; Schulz, S.; Höllt, V. Differential Expression of sst1, sst2A, and sst3Somatostatin Receptor Proteins in Low-Grade and High-Grade Astrocytomas. J. Neuropathol. Exp. Neurol. 2004, 63, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Schally, A.; Nagy, A. Chemotherapy targeted to cancers through tumoral hormone receptors. Trends Endocrinol. Metab. 2004, 15, 300–310. [Google Scholar] [CrossRef]
- Kiaris, H.; Schally, A.; Nagy, A.; Sun, B.; Szepeshazi, K.; Halmos, G. Regression of U-87 MG human glioblastomas in nude mice after treatment with a cytotoxic somatostatin analog AN-238. Clin. Cancer Res. 2000, 6, 709–717. [Google Scholar]
- Pinski, J.; Schally, A.; Halmos, G.; Szepeshazi, K.; Groot, K. Somatostatin analogues and bombesin/gastrin-releasing peptide antagonist RC-3095 inhibit the growth of human glioblastomas in vitro and in vivo. Cancer Res. 1994, 54, 5895–5901. [Google Scholar]
- Uphoff, C.C.; Drexler, H.G. Detection of Mycoplasma Contaminations. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]





| Parameters | Control Patients | Anaplastic Astrocytoma | Glioblastoma |
|---|---|---|---|
| Patients (n) | 8 | 9 | 47 |
| Gender (M/F) | 2(25%)/6(75%) | 3(33.3%)/6(66.7%) | 19(40.4%)/28(59.6%) |
| Age at surgical intervention (mean ± desvest) | 44.13 ± 4.32 | 51.9 ± 13.0 | 56.9 ± 13.2 |
| % Ki67 (mean ± desvest) | - | 15.2 ± 3.2% | 28 ± 14.6% |
| % of TP53 positive | - | 87.5% | 81.8% |
| % of IDH1 positive | - | 33% | 11.6% |
| % of recurrent tumors | - | 0% | 18% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentes-Fayos, A.C.; G-García, M.E.; Pérez-Gómez, J.M.; Peel, A.; Blanco-Acevedo, C.; Solivera, J.; Ibáñez-Costa, A.; Gahete, M.D.; Castaño, J.P.; Luque, R.M. Somatostatin Receptor Splicing Variant sst5TMD4 Overexpression in Glioblastoma Is Associated with Poor Survival, Increased Aggressiveness Features, and Somatostatin Analogs Resistance. Int. J. Mol. Sci. 2022, 23, 1143. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23031143
Fuentes-Fayos AC, G-García ME, Pérez-Gómez JM, Peel A, Blanco-Acevedo C, Solivera J, Ibáñez-Costa A, Gahete MD, Castaño JP, Luque RM. Somatostatin Receptor Splicing Variant sst5TMD4 Overexpression in Glioblastoma Is Associated with Poor Survival, Increased Aggressiveness Features, and Somatostatin Analogs Resistance. International Journal of Molecular Sciences. 2022; 23(3):1143. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23031143
Chicago/Turabian StyleFuentes-Fayos, Antonio C., Miguel E. G-García, Jesús M. Pérez-Gómez, Annabel Peel, Cristóbal Blanco-Acevedo, Juan Solivera, Alejandro Ibáñez-Costa, Manuel D. Gahete, Justo P. Castaño, and Raúl M. Luque. 2022. "Somatostatin Receptor Splicing Variant sst5TMD4 Overexpression in Glioblastoma Is Associated with Poor Survival, Increased Aggressiveness Features, and Somatostatin Analogs Resistance" International Journal of Molecular Sciences 23, no. 3: 1143. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23031143