In Vitro Metabolism of Dibenzo[a,l]pyrene, 2-Chlorodibenzo [a,l]pyrene and 10-Chlorodibenzo[a,l]pyrene - Effects of Chloro Substitution
Abstract
:Introduction
Materials and Methods
Materials
Metabolism of DB[a,l]P, 2-Cl-DB[a,l]P and 10-Cl-DB[a,l]P by rat liver microsomes
Mutagenicity assays
Physiochemical properties of metabolites
Results
HPLC separation and identification of metabolites
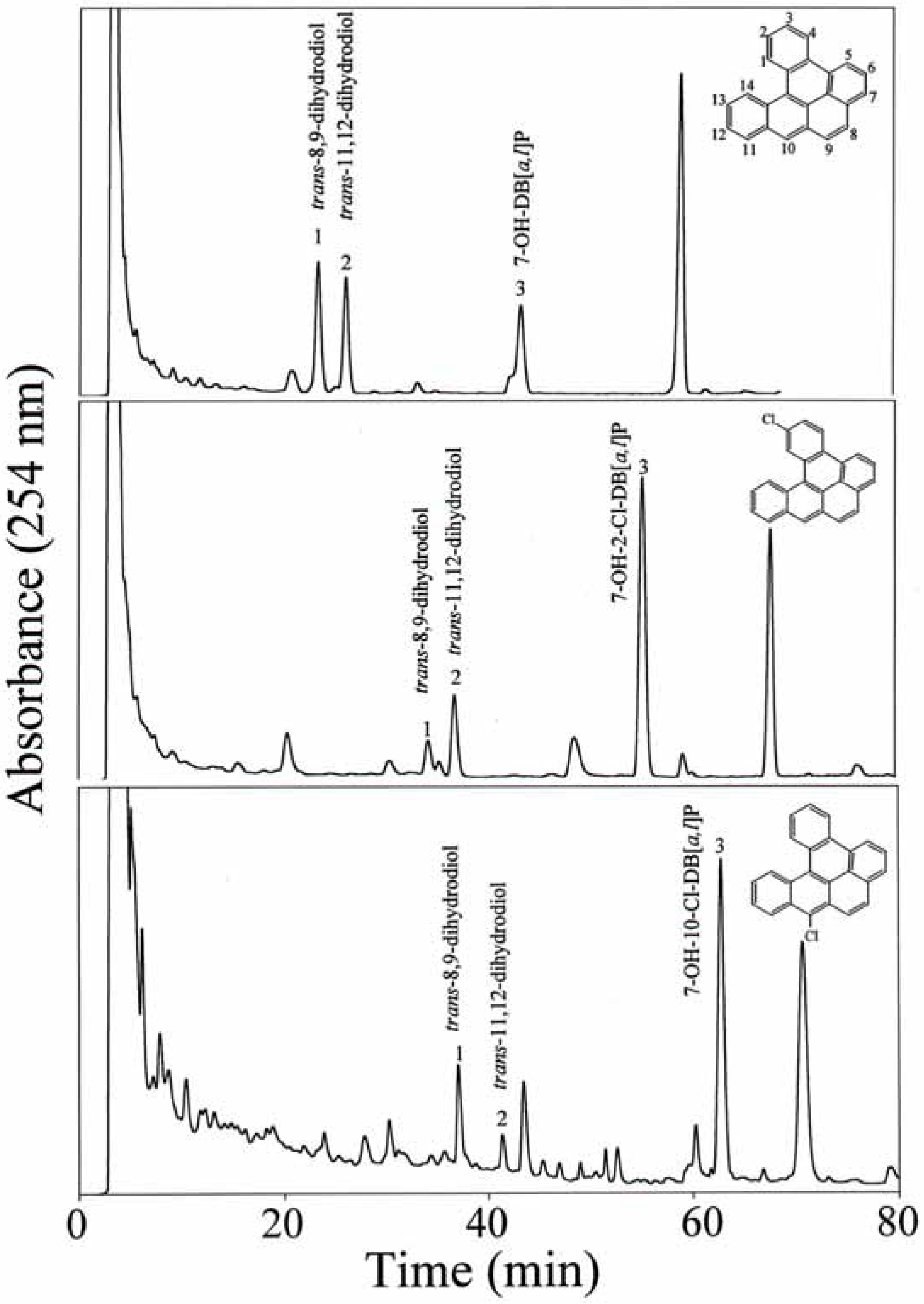
Quantification of metabolite formation
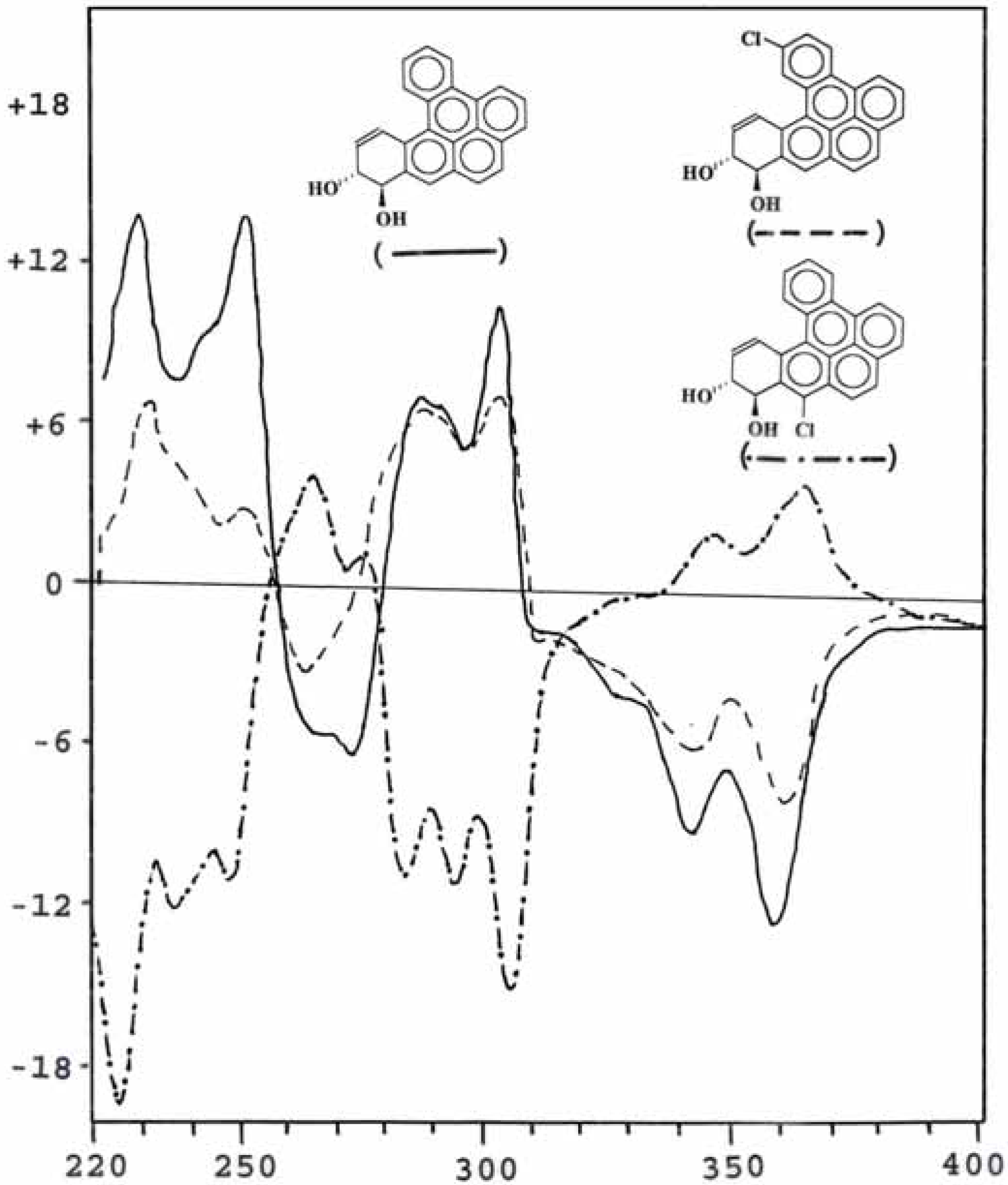
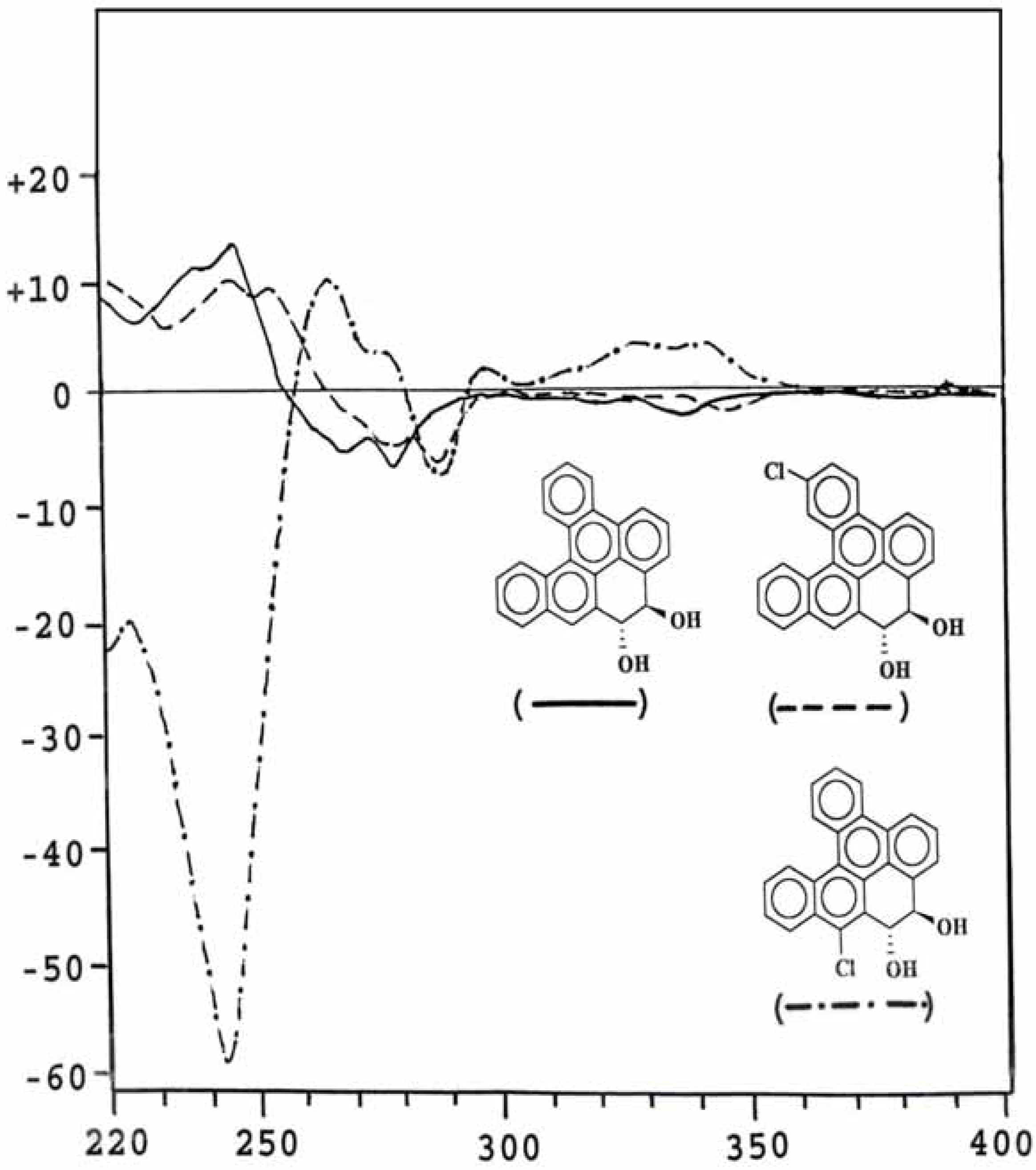
Conformation analysis of the trans-8,9-dihydrodiol metabolites
Optical property of metabolites
Mutagenicity of DB[a,l]P, 2-Cl-DB[a,l]P, and 10-Cl-DB[a,l]P
| Compound | Dose μg/plate | Revertants/plate (mean ± S.D.) |
|---|---|---|
| DMSO | 280 ±76 | |
| Benzo[a]pyrene | 1 μg | 1049 ± 112 |
| DB[a,l]P | 0.2 μg | 591 ± 89 |
| 2-Cl-DB[a,l]P | 1 μg | 353 ± 55 |
| 10-Cl-DB[a,l]P | 25 μg | 380 ± 48 |
Discussion

Acknowledgments
References
- Sovocool, G. W.; Mitchum, R. K.; Tondeur, Y.; Munslow, W. D.; Vonnahme, T. V.; Donnelly, J. R. Biomed. Environ. Mass Spectrom. 1988, 15, 669.
- Haglund, P.; Alsberg, T.; Bergman, A.; Jansson, B. Chemosphere 1987, 16, 2441.
- Nilsson, U. L.; Ostman, C. E. Environ. Sci. Technol. 1993, 27, 1826. [Google Scholar]
- Lofroth, G.; Nilsson, L.; Agurell, E.; Sugiyama, T. Mutat. Res. 1985, 155, 91–94.
- Fu, P. P.; Von Tungeln, L. S.; Chou, M. W. Mol. Pharmacol 1985, 28, 62.
- Fu, P. P.; Von Tungeln, L. S.; Chiu, L-H.; Own, Z. Y. Environ. Carcinogen. Ecotoxicol. Rev. 1999, C17(2), 71–109.
- Fu, P. P.; Von Tungeln, L. S.; Zhan, D.-J. Cancer Lett. 1996, 101, 37–42.
- Cavalieri, E. L.; Rogan, E. G.; Higginbotham, S.; Cremonesi, P.; Salmasi, S. J. Cancer Res. Clin. Oncol. 1989, 115, 67–72.
- Cavalieri, E. L.; Higginbotham, S.; Ramakrishna, N. V. S.; Devanesan, P. D.; Todorovic, R.; Rogan, E. G.; Salmasi, S. Carcinogenesis 1991, 12, 1939.
- Higginbotham, S.; Ramakishna, N. V. S.; Johansson, S. L.; Rogan, E. G.; Cavalieri, E. L. Carcinogenesis 1993, 14, 875.
- Devanesan, P. D.; Cremonesi, P.; Nunnally, J. E.; Rogan, E. G.; Cavalieri, E. L. Chem. Res. Toxicol. 1990, 3, 580.
- Masuda, Y.; Kagawa, R. Chem. Pharm. Bull. 1972, 20, 2736.
- Carruthers, W. J. Chem. Soc. 1967, 1525.
- Luch, A.; Glatt, H.; Platt, K. L.; Oesch, F.; Seidel, A. Carcinogenesis 1994, 15, 2507.
- Gill, H. S.; Kole, P. L.; Wiley, J. C.; Li, K.-M.; Higginbotham, S.; Rogan, E. C.; Cavalieri, E. L. Carcinogenesis 1994, 15, 2455.
- Amin, S.; Krzeminski, J.; Rivenson, A.; Kurtzke, C.; Hecht, S. S.; El-Bayoumy, K. Carcinogenesis 1995, 16, 1971.
- Luch, A.; Friesel, H.; Seidel, A.; Platt, K. L. Cancer Lett. 1999, 136, 119–128.
- Fu, P. P.; Von Tungeln, L. S.; Unruh, L. E.; Ni, Y. C.; Chou, M. W. Carcinogenesis 1991, 12, 371.
- Lowry, O. H.; Rosebrough, N. J.; Farr, A. L.; Randall, R. J. J. Biol. Chem. 1951, 193, 265.
- Vingiello, F. A.; Yanez, J.; Campbell, J. A. J. Org. Chem. 1971, 36, 2053. [CrossRef]
- Own, Z.Y.; Chiu, L.-H.; Von Tungeln, L.S.; Deck, J.; Vingiello, F. A.; Fu, P. P. Polycyclic Aromatic Compounds 1996, 11, 333–340.
- Maron, D. M.; Ames, B. N. Mutation Res. 1983, 113, 173–215.
- Fu, P. P.; Yang, S. K. Carcinogenesis 1983, 4, 979.
- Fu, P. P.; Yang, S. K. Biochem. Biophys. Res. Commun. 1983, 123.
- Yang, S. K.; Chou M. W.; Fu, P. P. in B. Pullman, P. O. P. Ts'o and H. Gelboin (editors), Carcinogenesis: Fundamental Mechanisms and Environmental Effects D. Reidel Publishing Co., Dordecht-Holland, 1980, p 143.
- Zacharias, D. E.; Glusker, J. P.; Fu, P. P.; Harvey, R. G. J. Am. Chem. Soc. 1979, 101, 4043. [CrossRef]
© 2002 by MDPI (http://www.mdpi.org).
Share and Cite
Wu, Y.-S.; Fang, G.-C.; Moody, J.; Von Tungeln, L.S.; Fu, P.P.; Hwang, H.-M.; Yu, H. In Vitro Metabolism of Dibenzo[a,l]pyrene, 2-Chlorodibenzo [a,l]pyrene and 10-Chlorodibenzo[a,l]pyrene - Effects of Chloro Substitution. Int. J. Mol. Sci. 2002, 3, 1008-1018. https://0-doi-org.brum.beds.ac.uk/10.3390/i3091008
Wu Y-S, Fang G-C, Moody J, Von Tungeln LS, Fu PP, Hwang H-M, Yu H. In Vitro Metabolism of Dibenzo[a,l]pyrene, 2-Chlorodibenzo [a,l]pyrene and 10-Chlorodibenzo[a,l]pyrene - Effects of Chloro Substitution. International Journal of Molecular Sciences. 2002; 3(9):1008-1018. https://0-doi-org.brum.beds.ac.uk/10.3390/i3091008
Chicago/Turabian StyleWu, Yuh-Shen, Guor-Cheng Fang, Joanna Moody, Linda S. Von Tungeln, Peter P. Fu, Huey-Min Hwang, and Hongtao Yu. 2002. "In Vitro Metabolism of Dibenzo[a,l]pyrene, 2-Chlorodibenzo [a,l]pyrene and 10-Chlorodibenzo[a,l]pyrene - Effects of Chloro Substitution" International Journal of Molecular Sciences 3, no. 9: 1008-1018. https://0-doi-org.brum.beds.ac.uk/10.3390/i3091008




