1. Introduction
Utilization of cellulose requires that microorganisms have cell-free cellulases or have such enzymes located on the outside of the cells. Also an enzyme found in a growing culture could be either secreted or passively released as a result of cell lysis [
1], making study of cellulolytic enzymes rather complicated. Cellulases occur in solutions in the form of components of different specificities which are often difficult to separate [
2]. In addition, assays with cellulose as a substrate cannot be obtained with high sensitivity; therefore carboxymethylcellulose (CMC) is often used as substrates to measure cellulase activity in bacteria [
3].
The cellulase location of most organisms has been studied.
Pseudomonas fluorescens has been found to have cell-bound and cell-free cellulase in varying proportions depending on the carbon source for growth [
4]. Chang [
5], reported in a study with
Cytophaga species WTHC 2421 (ATCC 29474), that cellulases were found in the soluble portion of the cell, primarily the periplasm and the cytoplasm. Preliminary data from our laboratory suggested a different mechanism by
Cytophaga hutchinsonii. A combined hydrolysis process was noticed, where part of the activity was cell-bound. Hence further investigation of the location of the cellulase in this species was conducted. This study gave additional insight into the mechanism of cellulose hydrolysis by
Cytophaga hutchinsonii.
Cytophaga are gram-negative bacteria. They are rod-shaped, but specific strains differ in diameter and length. Organisms of this group are ubiquitous and abundant in the environment. Their main habitats are soils and decaying plant material. They are found in freshwater environments, in estuaries and sediments. They are also common in sewage treatment plants.
Cytophaga have been shown to be the major cellulose degraders in some Canadian lakes [
6]. A few species have been isolated from the oral cavity of humans where they appear to occasionally cause septicemias [
7]. More importantly, the major role of
Cytophaga cells in the environment is the mineralization of organic matter.
Cellulose utilization generally proceeds via organisms that are either aerobic or anaerobic, but not both. In addition, there is a distinct difference in cellulolytic strategy between the aerobic and anaerobic groups. Anaerobes degrade cellulose primarily via complexed cellulase systems, cellulosome [
8]. Most anaerobic species that utilize cellulose do not release measurable amounts of extracellular cellulase. They have their cellulases localized directly on the surface of the cell. Aerobic cellulose degraders utilize cellulose through the production of substantial amounts of extracellular cellulase enzymes that are freely recoverable from culture supernatants [
9]. While many aerobic bacteria adhere to cellulose, physical contact between cells and cellulose does not appear to be necessary for cellulose hydrolysis [
10]. In the study presented here, we evaluated the cellulase system of the aerobic bacteria
Cytophaga hutchinsonii by determining the location, formation and biosynthetic regulation of its cellulase enzymes.
2. Methodology
Preliminary studies indicated that the production of cellulases can be affected by the growth media [
11]. Therefore for this experiment, colonies of
C. hutchinsonii scraped from culture plates were grown on minimal media supplemented with different carbon sources: glucose, cellobiose, cellulose, starch and xylan (Tab. 1). They were incubated in 250 ml flasks at an optimum growth temperature of 26°C with shaking. Ten ml of media including 10μl of crudem cellulase enzyme (from overnight culture on LB) were added to each flask and incubated with the respective carbon source. Samples were taken twice a day with a 9-hour-interval for a period of 7 days. The whole flask was removed at intervals and stored at -70°C for sugar and protein analysis. The experiment was repeated several times and each time with 3 replicates.
The different cellular fractions for the crude cellulase enzyme were prepared in Wilson’s laboratory at Cornell University's Department of Molecular Biology and Genetics, as illustrated in Fig 1. For the sugar and protein, modified procedures from Wilson’s lab [
11] were followed: 100 ml of culture taken from the stored 250ml flaks were centrifuged at maximum rpm (13,500 rpm). Protease inhibitor (PMSF) was added to the supernatant at a concentration of 0.1 mM. The resulting pellet was shocked by resuspension in 10ml NEST buffer. The supernatant was stored for further analysis after addition of 0.1 mM of PMSF. The shocked pellet was resuspended in 10 ml of 10 mM NaKPi (pH 7.4) and run through the French Press, to get the cytoplasmic fluid. The cell extract was treated with 0.1 mM of PMSF. The pellet was kept frozen at -70°C after addition of 1mM of PMSF. A portion of the pellet was treated with 0.5% Triton X-100 and analyzed for the formation of enzyme-trapped-vesicles. The different fractions were assayed for CMC activity [
3].
CMCase was assayed in terms of CMC-saccharifying activity. The latter was performed according to the method of Miller
et. al [
12], with three replicates and slight modification. The substrate contained 0.5% CMC (Sigma) in 0.1M sodium potassium phosphate buffer at pH 7.4. 400 μl of substrate was mixed with 0, 10, 20, 50 μl of crude cellulase enzyme and incubated at 30°C overnight. The reaction mixture was mixed with 200 μl of dinitrosalicylic acid (DNS) and heated in a boiling bath for 15 min. Each tube contained 2% CMC, 200 μl of the sugar to be tested, and incremental amounts of crude enzyme. The final volume in each tube was 800 μl before the addition of DNS reagent. The optical density of the mixture was determined at 660 nm. One unit of CMCase activity was defined as the amount of enzyme required to cause a 5% substrate digestion [
3]. For protein assay the method of Lowry
et. al. [
13] was used with bovine serum albumin (BSA) as the standard protein [
13]. A set of control tubes was prepared for both the protein and the sugars. One unit of specific activity of CMCase was defined as 1 mg glucose equivalent produced/mg protein per hour. In addition, cell adhesion was determined by microscopic observation. A compound microscope (MEIJI ML5000 Series) with binocular formats was used to visualize the cellulose structures. For our particular purpose, a magnification of 40X was sufficient. The observations were digitally captured and further analyzed (
Figure 3).
Figure 1.
Procedure utilized for physical and chemical separation of various cell fractions of Cytophaga hutchinsonii
Figure 1.
Procedure utilized for physical and chemical separation of various cell fractions of Cytophaga hutchinsonii
*Cell fractions were assayed separately for cellulase activity. DNS and Lowry Protein assays were run overnight at 30 ºC. Centrifugation was performed at 13,000 rpm for 10 min. Cells were shocked by transferring them from a low to a high concentration of NEST solution. French Press was used to turn the cells inside out, releasing thereby their cytoplasmic content.
3. Results and Discussion
The effects of sugars on CMCase activity are presented in
Table 1. The total and specific activities increased after the cells were plasmalyzed by lysozyme. For total activity, 1 unit of cellulose activity was expressed as the amount of protein that released reducing sugars equivalent to 1 µg of glucose per minute under assay conditions. Specific activity was expressed as unit of enzyme activity per mg of protein. Almost seventy percent of this activity was in the periplasm (
Table 2). The sum of the total activity of the periplasm and the broken spheroplasts was 3 times the total activity of the broken cell mixture which contained periplasm and the whole spheroplasts. Based on these findings, concurred also by Chang [
5], one can conclude that the cellulase enzyme of
C. hutchinsonii is located in the soluble portion of the cell, the periplasm.
The CMCase activity of various parts of the bacterial cell grown on different carbon sources is presented in
Table 1. The CMCase activity was slightly inhibited by the presence of glucose and cellobiose, and increased by the presence of cellulose. Little CMC-hydrolyzing enzyme was detected in the cell-free medium of cultures growing on glucose and cellobiose. Cultures grown on xylan formed at least 3 times less CMC-hydrolyzing enzymes as did cultures on cellulose. These results paralleled those of Lynd
et. al. [
14], where they found that, labile substrates, such as glucose or cellobiose acted as cellulase suppressors and cellulose as enzyme inducers.
Treatment of
C. hutchinsonii cells with the non-ionic detergent 0.5% Triton X100 caused rapid cell lysis and partial dissolution of remaining cell walls. When suspensions of bacteria were assayed for CMC-hydrolyzing enzyme before and after such treatment, no major increases in activity were observed with cells harvested from cultures grown on cellulose (Data not shown). The hypothesis that the formation of vesicles after The French Press treatment might have trapped some of the enzymes inside the cells, could not be proven correct, as was the case in Berg’s [
15] studies. All the cellulase enzymes seem to have been released into the cytoplasm.
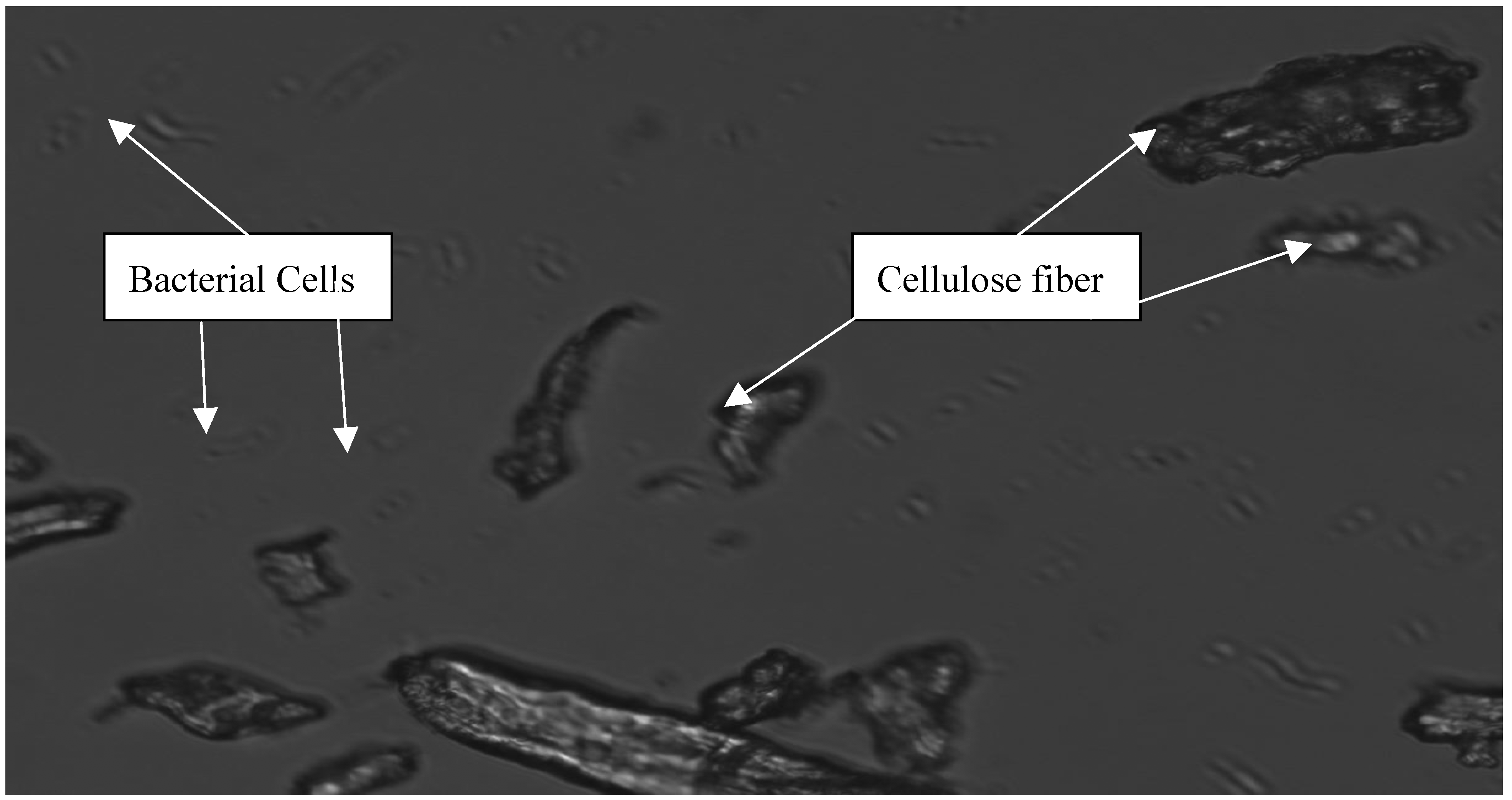
When cellulose fiber is examined under the microscope, it is clear that the microorganisms do not adhere to the cellulosic material very tightly (Fig. 3). Experiments conducted in the lab with other microorganisms, have also shown that if the bacteria cannot adhere to the cellulosic material, then cellulose will not be degraded efficiently [
16]. For these reasons it is believed that the rate and extent to which microorganisms adhere to cellulose is important for the digestibility of the material, and therefore the enzyme activity. Cell adhesion does not appear to be necessary in cellulose hydrolysis by
Cytophaga hutchinsonii.
Table 1.
Total and specific activities of various cell fractions separated from cells of Cytophaga Hutchinsonii grown on different carbon sources at 30°C.
Table 1.
Total and specific activities of various cell fractions separated from cells of Cytophaga Hutchinsonii grown on different carbon sources at 30°C.
| CARBON SOURCE | a TOTAL ACTIVITY (μmol reducing sugars/min per ml of culture) | b SPECIFIC ACTIVITY(total sugar/mg protein) |
|---|
| CELL FRACTIONS |
|---|
| WHOLE CELL | SUPER NATANT | SHOCK FLUID | CYTOPLASM | PELLET | WHOLE CELLS | SUPER NATANT | SHOCK FLUID | CYTOPLASM | PELLET |
|---|
| GLUCOSE | 0.17 | 0.34 | 0.04 | 0.12 | 0.09 | 15.45 | 24.29 | 3.08 | 7.06 | 8.18 |
| CELLOBIOSE | 0.21 | 0.41 | 0.10 | 0.23 | 0.12 | 7.00 | 15.19 | 4.00 | 12.11 | 10.00 |
| CELLULOSE | 0.72 | 1.53 | 0.21 | 0.37 | 0.32 | 12.86 | 61.2 | 16.15 | 30.83 | 24.62 |
| XYLAN | 0.26 | 0.52 | 0.09 | 0.23 | 0.18 | 7.65 | 14.05 | 3.46 | 10.00 | 10.59 |
| STARCH | 0.19 | 0.37 | 0.12 | 0.35 | 0.23 | 7.04 | 19.47 | 3.87 | 14.00 | 6.39 |
Table 2.
Hydrolysis assay of different cell fractions separated from C. hutchinsonii cells grown on solka flok (cellulose) as the carbon source.
Table 2.
Hydrolysis assay of different cell fractions separated from C. hutchinsonii cells grown on solka flok (cellulose) as the carbon source.
| **OPTICAL DENSITY (OD) | CULTURE MEDIUM (μl) | OD/ml Culture | ***ENZYME ACTIVITY (%) | *OD/min – ml |
|---|
| WHOLE CELLS (w) | 0.72 | 10 | 72 | 32% | 0.090 |
| CYTOPLASMIC FLUID (C) | 0.37 | 20 | 9 | | 0.020 |
| PELLET SUSPENSION (P) | 0.32 | 20 | 16 | | 0.020 |
| SUPERNATANT (S) | 1.53 | 153 | 16 | 68% | 0.200 |
| SHOCK FLUID (F) | 0.21 | 50 | 2.6 | | 0.060 |
| C + P+ F | 0.57 | | 25 | | |
| W + S | | | | | |
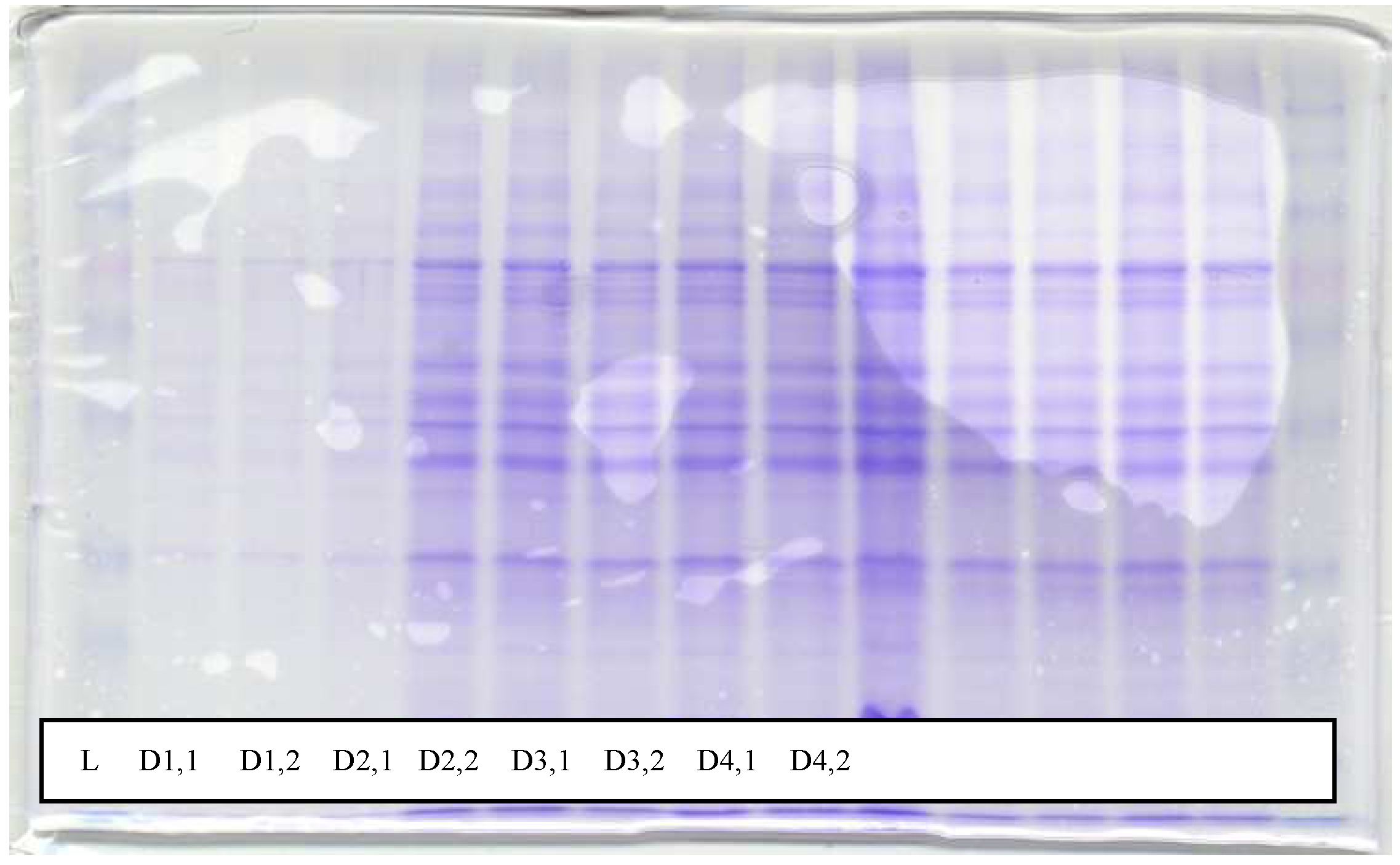
Figure 2.
SDS gel showing overtime distribution of protein in the supernatant over a period of 7 days. L is the loading buffer used on a 12.5% gel. D1,1 and D1,2 represent morning and afternoon samples on the first day, D2,1 and D2,2 represent morning and afternoon samples on the second day, D3,1 and D3,2 represent morning and afternoon samples on the third day...and so on. Since the microorganisms reached their log phase after the 4th day, the continuous protein release into the culture solution (D4 to D7) was probably due to cell lysis.
Figure 2.
SDS gel showing overtime distribution of protein in the supernatant over a period of 7 days. L is the loading buffer used on a 12.5% gel. D1,1 and D1,2 represent morning and afternoon samples on the first day, D2,1 and D2,2 represent morning and afternoon samples on the second day, D3,1 and D3,2 represent morning and afternoon samples on the third day...and so on. Since the microorganisms reached their log phase after the 4th day, the continuous protein release into the culture solution (D4 to D7) was probably due to cell lysis.
Figure 3.
Microscopic observation of C. hutchinsonii grown on cellulose revealed no level of cell adhesion by this microorganism to the cellulose fibers. Picture taken after 96h incubation when most of the enzyme activity was measured.
Figure 3.
Microscopic observation of C. hutchinsonii grown on cellulose revealed no level of cell adhesion by this microorganism to the cellulose fibers. Picture taken after 96h incubation when most of the enzyme activity was measured.
Conclusions
Cellulase systems have been studied extensively in other organisms. Cottrell et al. [
17] presented further evidence of the importance of
Cytophaga-like bacteria in the degradation of protein in high-molecular-weight dissolved organic matter. Karlsson et al [
18] suggested that Xyn10A and Man26A represent a novel type of module that mediates cell attachment in proteins originating from members of the phylum
Bacteroidetes. Most of the
T. fusca cellulases have been cloned, expressed and characterized [
12]. Lynd [
23] provided an extensive review on studied cellulases from aerobic and anaerobic microorganisms. From these studies and others, we found that bacterial enzymes may be restricted to particular regions in the cells or actively secreted into the surrounding medium [
1]. Enzymes which attack degraded cellulose such as CMC may be measured in terms of CMC-liquefying activity or CMC-saccharifying activity. Emert
et. al. [
19] proposed that the former was only exhibiting the activity of endo-ß-1, 4-glucanase and the latter the activity of exo-ß-1,4-glucanase. In this experiment both were assayed in the same crude enzyme preparations. The results indicated that most of the CMC-saccharifying enzyme was soluble. It is not known whether other enzymes were involved in the cellulose reaction and affected the outcome of the assays. Enzymes capable of attacking the products of cellulose hydrolysis (glucose oxidase, hexokinase and glucose dehydrogenase) are all soluble [
20]. Therefore it is unlikely that they were present on the membrane and resulted in erroneously low results for CMC-saccharifying activity.
Catabolism of cellulose involves both enzymatic depolymerization of insoluble cellulose and cellular utilization of the hydrolytic products. There are two primary strategies for utilizing crystalline cellulose. Aerobic bacteria and fungi do not adhere (or adhere only weakly) to cellulose, produce noncomplexed cellulases, and oxidize hydrolytic products to CO
2 and H
20. Anaerobic bacteria and fungi display a greater tendency (or in some cases a requirement) to adhere to cellulose; produce primarily complexed cellulases exemplified by the cellulosome organelle. For anaerobic bacterial species, adhesion of cells to cellulose is much more common, and for some species it appears to be a requirement for rapid and efficient cellulose hydrolysis [
21]. Hofsten and Berg [
22] proposed that cellulases are more effective when cell bound because they are in high concentration and are favorably aligned with the substrate. Cellulose hydrolysis requires prior binding of enzymes to cellulose, either as an enzyme-substrate binary complex or as a cellulose-enzyme-microbe (CEM) ternary complex [
23]. Some cells with noncomplexed cellulase systems (e.g., Cellulomonas sp. strain NRCC 2406) show a tendency to adhere to cellulose, although such contact does not appear to be necessary for cellulose utilization, as in the case of
C. hutchinsonii (Fig 3).
It has been have shown that
Bacteroides spp., a distantly related species of
C. hutchinsonii, use a different strategy [
19]. Polysaccharides are first bound to the cell surface and are then translocated across the outer membrane into the periplasm. The degradative enzymes, most of which appear to be located in the periplasm, then break down the polysaccharides and the resulting products are transported across the cytoplasmic membrane into the cytoplasm. This type of strategy makes sense in a highly competitive ecosystem such as that found in the human colon than the extracellular enzyme strategy, because it helps to prevent products of polysaccharide breakdown from being lost to competing bacteria. In this study, we presented evidence that celllases from the aerobic soil bacteria,
C. hutchinsonii, are predominantly cell-free during active growth on cellulose, which acts as an enzyme inducer, while glucose and cellobiose seems to repress cellulase formation. In the light of these findings, we hypothesize that
C. hutchinsonii attack polysaccharides by secreting extracellular enzymes, which degrade the polysaccharide to subunits small enough to be transported into the cytoplasm. However, this hypothesis remains to be tested.