Studies on the Interaction between Zinc-Hydroxybenzoite Complex and Genomic DNA
Abstract
:1. Introduction
2. Results and Discussion
2.1 Interaction between [Zn(H20)6] (p-HO-C6H4COO)2. 2H20 complex and genomic DNA

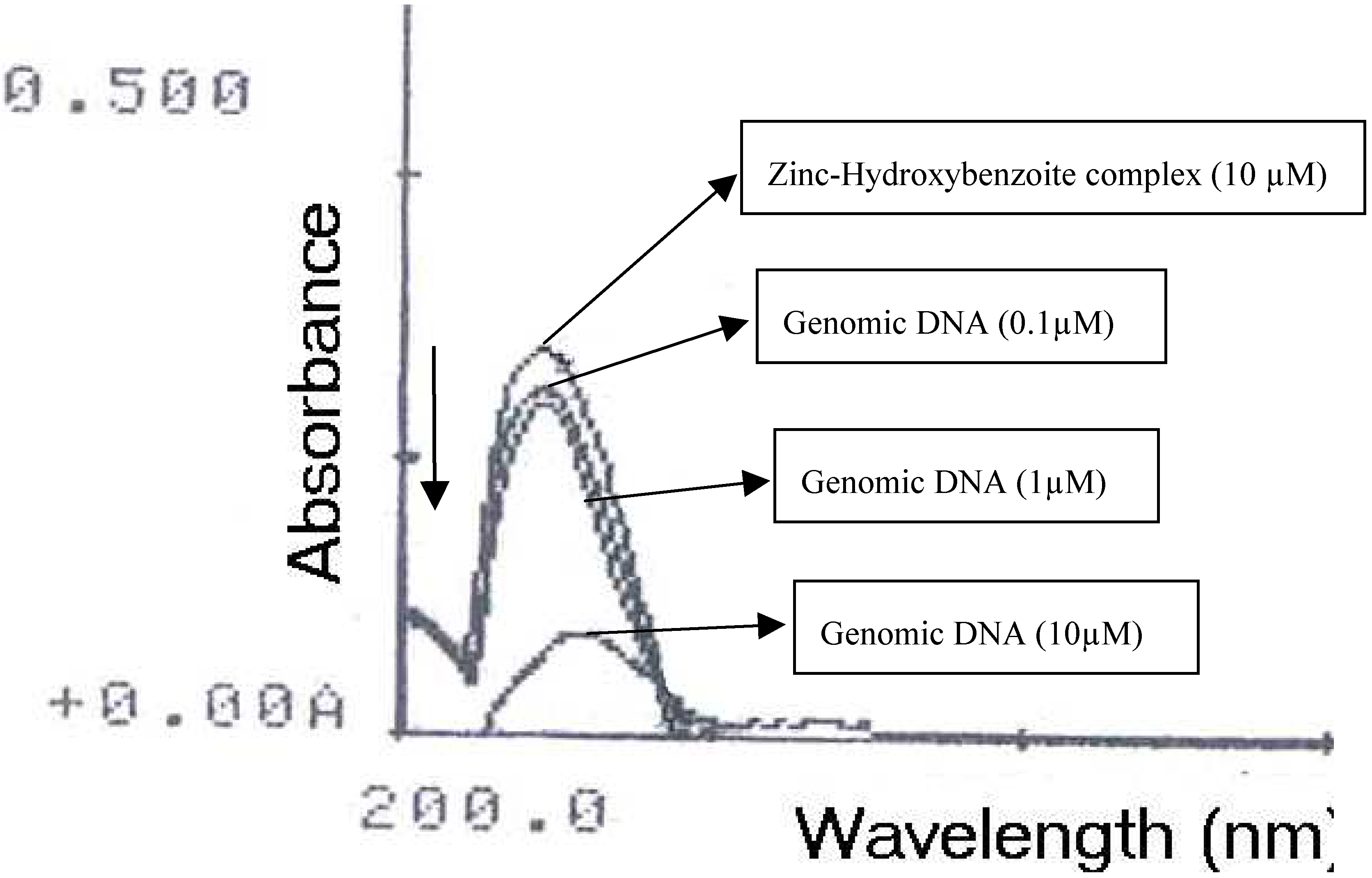
2.2 Conclusion
3. Experimental Section
3.1 Materials
3.2 Synthesis
3.3 Methods
3.3.1 [Zn(H20)6](p-HO-C6H4COO)2·2H20 Genomic DNA Binding
3.3.2 Gel Electrophoresis
3.3.3 Interaction between [Zn(H20)6](p-HO-C6H4COO)2·2H20 complex and genomic DNA
Acknowledgements
References and Notes
- Bray, T.M.; Bettger, W.J. The physiological role of zinc as an antioxidant. Free Radic. Biol. Med. 1990, 8, 281–291. [Google Scholar] [CrossRef]
- Powell, S.R. The antioxidant properties of zinc. J. Nutr. 2000, 130, 1447S–1454S. [Google Scholar]
- Dreosti, I.E. Zinc and the gene. Mutate. Res. 2001, 475, 161–167. [Google Scholar] [CrossRef]
- Cousins, R.J. A role of zinc in the regulation of gene expression. Proc. Nutr. Soc. 1998, 57, 307–311. [Google Scholar] [CrossRef]
- Falchuk, K.H. The molecular basis for the role of zinc in developmental biology. Mol. Cell Biochem. 1998, 188, 41–48. [Google Scholar] [CrossRef]
- Prasad, A.S. Zinc deficiency in humans: a neglected problem. J. Am. Coll. Nutr. 1998, 17, 542–543. [Google Scholar] [CrossRef]
- Prasad, A.S.; Kucuk, O. Zinc in cancer prevention. Cancer Metastasis Rev. 2002, 21, 291–295. [Google Scholar] [CrossRef]
- Prasad, A.S. Zinc deficiency. BMJ. 2003, 326, 409–410. [Google Scholar] [CrossRef]
- Erkkila, K.E.; Odom, D.T.; Barton, J.K. Recognition and reaction of metallointercalators with DNA. Chem. Rev. 1999, 99, 2777–2796. [Google Scholar] [CrossRef]
- Metcalfe, C.; Thomas, J.A. Kinetically inert transition metal complexes that reversibly bind to DNA. Chem. Soc. Rev. 2003, 32, 215–224. [Google Scholar] [CrossRef]
- Haq, I.; Lincoln, P.; Suh, D.; Norden, B.; Choedhry, B.Z.; Chaires, J.B. Interaction of Delta A- [Ru (PHEN)(2)DPPZ](2+) and Lambda-[Ru(Phen)(2)DPPZ](2+) WITH DNA - A Caloimetric and Equilibrium Binding Study. J. Am. Chem. Soc. 1995, 17, 4788–4796. [Google Scholar]
- Arturo, S.; Giampaolo, B.; Giuseppe, R.; Maria, L.G.; Salvatore, T. The interaction of native DNA with Iron (III)-N, N′-ethylene-bis (salicylideneiminato)-chloride. J. Inorg. Biochem. 2004, 98, 589–594. [Google Scholar]
- Maribel, N.; Efren, C.F.; Anßbal, S.; Mercedes, F.M.; Pedro, S.; Dwight, A.; Edgar, M. Design of copper DNA intercalators with leishmanicidal activity. J. Biol. Inorg. Chem. 2003, 8, 401–408. [Google Scholar]
- Catherine, H.; Marguerite, P.; Michael, R.; Heinz, G.; Stephanie, S.; Bernard, M. Preparation, characterization and crystal structures of manganese (II), iron (III) and copper (II) complexes of the bis[di-1,1-(2-pyridyl) ethyl] amine (BDPEA) ligand; evaluation of their DNA cleavage activities. J. Biol. Inorg. Chem. 2001, 6, 14–22. [Google Scholar] [CrossRef]
- Kumar, C.V.; Barton, J.K.; Turro, N.J. Photophysics of Ruthenium Complexes Bound to Double Helical DNA. J. Am. Chem. Soc. 1985, 107, 5518–5523. [Google Scholar] [CrossRef]
- Xu, H.; Zheng, K.C.; Deng, H.; Lin, L.J.; Zhang, Q.L.; Ji, L.N. Effects of ligand planarity on the interaction of polypyridyl Ru (II) complexes with DNA. Dalton. Trans. 2003, 3, 2260–2268. [Google Scholar]
- Mahadevan, S.; Palaniandavar, M. Electrochemical study of the enantioselective interaction of Tris(phen)Ru (II) with calf thymus DNA. Inorg. Chim. Acta. 1997, 254, 291–302. [Google Scholar]
- Xu, H.; Zheng, K.C.; Deng, H.; Lin, L.J.; Zhang, Q.L.; Ji, L.N. Interaction of Ru(II) Complex with Yeast tRNA Studied by Isothermal Clarometry. New J. Chem. 2003, 27, 1255–1263. [Google Scholar] [CrossRef]
- Mozaffar, A.; Elham, S.; Bijan, R.; Leila, H. Thermodynamic and spectroscopic study on the binding of cationic Zn(II) and Co(II) tetrapyridinoporphyrazines to calf thymus DNA: the role of the central metal in binding parameters. New J. Chem. 2004, 28, 1227–1234. [Google Scholar] [CrossRef]
- Chaires, J.B. Energetics of drug-DNA binding. Biopolymers 1998, 44, 201–215. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, C.-S.; Bu, X.-H.; Yang, M. Synthesis, crystal structure, cytotoxic activity and DNA-binding properties of the copper (II) and zinc (II) complexes with 1-[3-(2-pyridyl) pyrazol-1-ylmethyl] naphthalene. J. Inorg. Biochem. 2005, 99, 1119–1125. [Google Scholar] [CrossRef]
- Shnulin, A.N.; Nadzhatov, G.N.; Amiraslanov, I.R.; Usubaliyev, B.T.; Mamedov, K.S. Diaquabis (N, N-diethylnicotinamide-N1)bis(2-hydroxybenzoato-O)cobalt(II). Koordinasionnaya Khimiya 1981, 7, 1409. [Google Scholar]
- Wang, X-L.; Chao, H.; Li, H.; Hang, X-L.; Liu, Y-J.; Tan, L-F.; Ji, L-N. DNA Interactions of Cobalt (III) Mixed-Polypridyl Complexes Containing Asymmetric Ligands. J. Inorg. Biochem. 2004, 98, 1143–1150. [Google Scholar] [CrossRef]
- Hossaian, Z.; Huq, F. Studies on the Interaction Between Ag+ and DNA. J. Inorg. Biochem. 2002, 91, 398–404. [Google Scholar] [CrossRef]
© 2006 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Arslantas, A.; Devrim, A.K.; Kaya, N.; Necefoglu, H. Studies on the Interaction between Zinc-Hydroxybenzoite Complex and Genomic DNA. Int. J. Mol. Sci. 2006, 7, 111-118. https://0-doi-org.brum.beds.ac.uk/10.3390/i7040111
Arslantas A, Devrim AK, Kaya N, Necefoglu H. Studies on the Interaction between Zinc-Hydroxybenzoite Complex and Genomic DNA. International Journal of Molecular Sciences. 2006; 7(4):111-118. https://0-doi-org.brum.beds.ac.uk/10.3390/i7040111
Chicago/Turabian StyleArslantas, Ali, A. Kadir Devrim, Neceti Kaya, and Hacali Necefoglu. 2006. "Studies on the Interaction between Zinc-Hydroxybenzoite Complex and Genomic DNA" International Journal of Molecular Sciences 7, no. 4: 111-118. https://0-doi-org.brum.beds.ac.uk/10.3390/i7040111



