TFIP11 Interacts with mDEAH9, an RNA Helicase Involved in Spliceosome Disassembly
Abstract
:1. Introduction
2. Experimental Section
2.1. Plasmid Constructs
2.2. Immunoprecipitation and Western Blot
2.3. Transfection, Immunocytochemistry and Imaging
3. Results
3.1. The localization of the transfected TFIP11-EGFP fusion protein mimics that of endogenous TFIP11
3.2. The G-patch of TFIP11 is not essential for the characteristic nuclear distribution of TFIP11
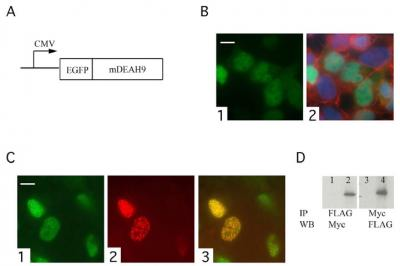
3.3. TFIP11 colocalizes with mDEAH9 in vivo, and interacts with mDEAH9 in vitro
4. Discussion
5. Conclusions
Acknowledgments
- The gene locus for mDEAH9 is identified as Dhx15, and two alternatively spliced isoforms are recognized for this gene. Previously published cDNA sequence data for mDEAH9 [10] (GenBank accession #AF017153) ended the coding region with “ATC AAG CCA GAA TGT TGG TGA” (stop codon underlined). The GenBank entry for Dhx15 isoform 2 (NM_007839) reads “ATC AAG CCA GAA TGG TTG GTG etc.” at this same location; and includes an additional G identified here in italics. This later GenBank entry recognizes a translated product for Dhx15 that includes an additional 39 amino acids at the C-terminus, when compared to mDEAH9. The mDEAH9 cDNA reported in this study was generated by RT-PCR using a reverse primer designed to the nucleotide sequence corresponding to the earlier reported C-terminus of mDEAH9 [10], thus coding for a product that equates to the N-terminal 756 amino acids of Dhx15 isoform 2.
References
- Zhou, Z; Licklider, LJ; Gygi, SP; Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 2002, 419, 182–185. [Google Scholar]
- Jurica, MS; Moore, MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 2003, 12, 5–14. [Google Scholar]
- Ji, Q; Huang, CH; Peng, J; Hashmi, S; Ye, T; Chen, Y. Characterization of STIP, a multi-domain nuclear protein, highly conserved in metazoans, and essential for embryogenesis in Caenorhabditis elegans. Exp. Cell Res 2007, 313, 1460–72. [Google Scholar]
- Wen, X; Lei, YP; Zhou, YL; Okamoto, CT; Snead, ML; Paine, ML. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11). Cell. Mol. Life Sci 2005, 62, 1038–1046. [Google Scholar]
- Boon, KL; Auchynnikava, T; Edwalds-Gilbert, G; Barrass, JD; Droop, AP; Dez, C; Beggs, JD. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell Biol 2006, 26, 6016–6023. [Google Scholar]
- Tsai, RT; Fu, RH; Yeh, FL; Tseng, CK; Lin, YC; Huang, YH; Cheng, SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 2005, 19, 2991–3003. [Google Scholar]
- Tsai, RT; Tseng, CK; Lee, PJ; Chen, HC; Fu, RH; Chang, KJ; Yeh, FL; Cheng, SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell Biol 2007, 27, 8027–8037. [Google Scholar]
- Pandit, S; Lynn, B; Rymond, BC. Inhibition of a spliceosome turnover pathway supresses splicing defects. Proc. Natl. Acad. Sci. USA 2006, 103, 13700–13705. [Google Scholar]
- Tanaka, N; Aronova, A; Schwer, B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 2007, 21, 2312–2325. [Google Scholar]
- Gee, S; Krauss, SW; Miller, E; Aoyagi, K; Arenas, J; Conboy, JG. Cloning of mDEAH9, a putative RNA helicase and mammalian homologue of Saccharomyces cerevisiae splicing factor Prp43. Proc. Natl. Acad. Sci. USA 1997, 94, 11803–11807. [Google Scholar]
- Stoss, O; Stoilov, P; Hartmann, AM; Nayler, O; Stamm, S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. Brain Res. Protoc 1999, 4, 383–394. [Google Scholar]
- Sambrook, J; Russell, DW. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, 2001. [Google Scholar]
- Aravind, L; Koonin, EV. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci 1999, 24, 342–344. [Google Scholar]
- Silverman, EJ; Maeda, A; Wei, J; Smith, P; Beggs, JD; Lin, RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell Biol 2004, 24, 10101–10110. [Google Scholar]


| mTFIPmut forward | 5′- CTACGTCCCTGCGCGTGCCCTGCGGAAGAACGCAC |
| mTFIPmut reverse | 5′- GTGCGTTCTTCCGCAGGGCACGCGCAGGGACGTAG |
| EGFP_mDEAH9 forward | 5′- GAATTCTATGTCCAAGAGGCATCGGTT |
| Myc_mDEAH9 forward | 5′- GAATTCTTATGTCCAAGAGGCATCGGTT |
| EGFP/Myc_mDEAH9 reverse | 5′- TCACCAACATTCTGGCTTGATA |
© 2008 by MDPI This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wen, X.; Tannukit, S.; Paine, M.L. TFIP11 Interacts with mDEAH9, an RNA Helicase Involved in Spliceosome Disassembly. Int. J. Mol. Sci. 2008, 9, 2105-2113. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms9112105
Wen X, Tannukit S, Paine ML. TFIP11 Interacts with mDEAH9, an RNA Helicase Involved in Spliceosome Disassembly. International Journal of Molecular Sciences. 2008; 9(11):2105-2113. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms9112105
Chicago/Turabian StyleWen, Xin, Sissada Tannukit, and Michael L. Paine. 2008. "TFIP11 Interacts with mDEAH9, an RNA Helicase Involved in Spliceosome Disassembly" International Journal of Molecular Sciences 9, no. 11: 2105-2113. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms9112105