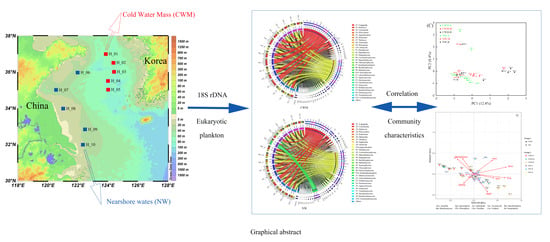
Characteristics of Eukaryotic Plankton Communities in the Cold Water Masses and Nearshore Waters of the South Yellow Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Analysis of Environmental Samples
2.2. Eukaryotic Plankton Community Characterization
2.3. Diversity Analysis and Statistical Analysis
3. Result
3.1. Sequencing Data of 18S rDNA
3.2. Structural Characteristics of the Eukaryotic Plankton Community
3.3. Changes in Composition and Diversity of Eukaryotic Plankton Community
3.4. Environmental Factor Data Analysis
3.5. Comparative Assessment of Microbial Biomarkers
3.6. Correlation Analysis between Environmental Factors and Eukaryotic Planktonic Community Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bigg, G.R.; Jickells, T.D.; Liss, P.S.; Osborn, T.J. The role of the oceans in climate. Int. J. Climatol. 2003, 23, 1127–1159. [Google Scholar] [CrossRef]
- Tatiana, A.R.; Ellen, O.L.; Virginia, A.E. Metapopulation structure in the planktonic diatom ditylum brightwellii (Bacillariophyceae). Protist 2009, 160, 111–121. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, S.; Zhao, R.; Zhu, L. Plankton community structure analysis and water quality bioassessment in Jiulong Lake. IOP Conf. Ser. Earth Environ. Sci. 2018, 199, 022031. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, C.; Jiang, Z.; Guo, S.; Sun, J. Seasonal variations in phytoplankton community structure in the Sanggou, Ailian, and Lidao Bays. J. Ocean Univ. China 2014, 13, 1012–1024. [Google Scholar] [CrossRef]
- Eiler, A.; Drakare, S.; Bertilsson, S.; Pernthaler, J.; Peura, S.; Rofner, C.; Lindström, E.S. Unveiling distribution patterns of freshwater phytoplankton by a next generation sequencing based approach. PLoS ONE 2013, 8, e53516. [Google Scholar] [CrossRef]
- Bianchi, F.; Acri, F.; Aubry, F.B.; Berton, A.; Boldrin, A.; Camatti, E.; Comaschi, A. Can plankton communities be considered as bio-indicators of water quality in the Lagoon of Venice? Mar. Pollut. Bull. 2003, 46, 964–971. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.; Zhang, X.; Wei, Y.; Qiu, L.; Wei, Z.; Li, F. Spatial heterogeneity in a deep artificial lake plankton community revealed by PCR-DGGE fingerprinting. Chin. J. Oceanol. Limnol. 2015, 33, 624–635. [Google Scholar] [CrossRef]
- Sun, J. Marinephytolankton and biological carbon sink. Acta Ecol. Sin. 2011, 31, 5372–5378. [Google Scholar]
- Charlson, R.J.; Lovelock, J.E.; Andreae, M.O.; Warren, S.G. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 1987, 326, 655–661. [Google Scholar] [CrossRef]
- Sathyendranath, S.; Gouveia, A.D.; Shetye, S.R. Biological control of surface temperature in the Arabian Sea. Nature 1991, 349, 54–56. [Google Scholar] [CrossRef]
- Sun, J. Geometric models for calculating cell biovolume and surface area for phytoplankton. J. Plankton Res. 2003, 25, 1331–1346. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.A.D.; Train, S.; Rodrigues, L.C. Phytoplankton assemblages in a Brazilian subtropical cascading reservoir system. Hydrobiologia 2005, 537, 99–109. [Google Scholar] [CrossRef]
- Warwick, R.M. The nematode/copepod ratio and its use in pollution ecology. Mar. Pollut. Bull. 1981, 12, 329–333. [Google Scholar] [CrossRef]
- Goh, T.K.; Hyde, K.D. Biodiversity of freshwater fungi. J. Ind. Microbiol. 1996, 17, 328–345. [Google Scholar] [CrossRef]
- Cudowski, A.; Pietryczuk, A.; Hauschild, T. Aquatic fungi in relation to the physical and chemical parameters of water quality in the Augustów Canal. Fungal Ecol. 2015, 13, 193–204. [Google Scholar] [CrossRef]
- Xiao, X.; Sogge, H.; Lagesen, K.; Tooming, K.A.; Jakobsen, K.S.; Rohrlack, T. Use of high throughput sequencing and light microscopy show contrasting results in a study of phytoplankton occurrence in a freshwater environment. PLoS ONE 2014, 9, e106510. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.S.; Hilary, G.M.; Julie, A.H.; David, M.W.; Susan, M.H.; Phillip, R.N.; Jesus, M.A.; Gerhard, J.H. Microbial diversity in the deep sea and the underexplored “Rare Biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [Green Version]
- Bucklin, A.; Frost, B.; Bradford, G.J.; Allen, L.; Copley, N. Molecular systematic and phylogenetic assessment of 34 calanoid copepod species of the Calanidae and Clausocalanidae. Mar. Biol. 2003, 142, 333–343. [Google Scholar] [CrossRef]
- Behnke, A.; Engel, M.; Christen, R.; Nebel, M.; Klein, R.R.; Stoeck, T. Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ. Microbiol. 2010, 13, 340–349. [Google Scholar] [CrossRef]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [Green Version]
- Zou, E.M.; Guo, B.H.; Tang, Y.X.; Lee, J.H.; Xiong, X.J.; Zeng, X.M. The hydrographic features and mixture and exchange of sea water in the southern Huanghai Sea in autumn. Acta Oceanol. Sinca 1999, 21, 12–21. [Google Scholar] [CrossRef]
- Xia, C.; Qiao, F.; Yang, Y.; Ma, J.; Yuan, Y. Three-dimensional structure of the summertime circulation in the Yellow Sea from a wave-tide-circulation coupled model. J. Geophys. Res. 2006, 111. [Google Scholar] [CrossRef]
- Lü, X.; Qiao, F.; Xia, C.; Wang, G.; Yuan, Y. Upwelling and surface cold patches in the Yellow Sea in summer: Effects of tidal mixing on the vertical circulation. Cont. Shelf Res. 2010, 30, 620–632. [Google Scholar] [CrossRef]
- Sun, S.; Wang, R.; Zhang, G.T.; Yang, P.; Zhang, F. A preliminary study on the over-summer strategy of Calanus Sinicus in the Yellow Sea. Oceanol. Limnol. Sinica 2002, 92–99. [Google Scholar]
- Oh, K.H.; Lee, S.; Song, K.M.; Lie, H.J.; Kim, Y.T. The temporal and spatial variability of the Yellow Sea Cold Water Mass in the southeastern Yellow Sea, 2009–2011. Acta Oceanol. Sin. 2013, 32, 1–10. [Google Scholar] [CrossRef]
- Jeffrey, S.W.; Humphrey, G.F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. und Physiol. der Pflanz. 1975, 167, 191–194. [Google Scholar] [CrossRef]
- Yuan, J.; Li, M.; Lin, S. An Improved DNA Extraction method for efficient and quantitative recovery of phytoplankton diversity in natural assemblages. PLoS ONE 2015, 10, e0133060. [Google Scholar] [CrossRef]
- Maral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M.; Langsley, G. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheik, C.S.; Mitchell, T.W.; Rizvi, F.Z.; Rehman, Y.; Faisal, M.; Hasnain, S.; Krumholz, L.R. Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS ONE 2010, 7, e40059. [Google Scholar] [CrossRef] [PubMed]
- Hajibabaei, M.; Shokralla, S.; Zhou, X.; Singer, G.A.C.; Baird, D.J. Environmental barcoding: A next-generation sequencing approach for biomonitoring applications using river benthos. PLoS ONE 2011, 6, e17497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.D.; Li, C.L. Classification of zooplankton communities in the adjacent waters of the Changjiang River estuary based on data from 1998 to 2011. Mar. Sci. 2018, 42, 38–47. [Google Scholar] [CrossRef]
- Liu, L.H. The Community Structure and Diversity Analysis of Phytoplankton in the Yellow Sea and the Chang Jiang Estuary Waters. Ph.D. Thesis, Ocean University of China, Shandong, China, 2007. (In Chinese). [Google Scholar] [CrossRef]
- Hamm, C.E.; Merkel, R.; Springer, O.; Jurkojc, P.; Maier, C.; Prechtel, K.; Smetacek, V. Architecture and material properties of diatom shells provide effective mechanical protection. Nature 2003, 421, 841–843. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yan, T.; Lin, J.; Yu, R.; Zhou, M. Detrimental impacts of the dinoflagellate Karenia mikimotoi in Fujian coastal waters on typical marine organisms. Harmful Algae 2017, 61, 1–12. [Google Scholar] [CrossRef]
- Tang, D.L.; Ni, I.H.; MüllerKarger, F.E.; Liu, Z.J. Analysis of annual and spatial patterns of CZCS-derived pigment concentration on the continental shelf of China. Cont. Shelf Res. 1998, 18, 1493–1515. [Google Scholar] [CrossRef]
- Ning, X.X.; Ji, L.; Wang, G.; Xia, B.X. Phytoplankton community in the nearshore waters of Laizhou Bay in 2009. Trans. Oceanol. Limnol. 2011, 3, 97–104. [Google Scholar] [CrossRef]
- Zuo, T.; Wang, R.; Chen, Y.Z.; Gao, S.W.; Wang, K. Net macro-zooplankton community classification on the shelf area of the East China Sea and the Yellow Sea in spring and autumn. Acta Ecol. Sin. 2005, 25. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, S.; Xu, Z. Marine Zooplankton Biology; China Ocean Press: Beijing, China, 1984; pp. 139–491. [Google Scholar]
- Uye, S. Temperature-dependent development and growth of Calanus sinicus (Copepoda: Calanoida) in the laboratory. Hydrobiologia 1988, 167, 285–293. [Google Scholar] [CrossRef]
- Huo, Y.Z. Study on the Zooplankton Functional Groups in the Yellow Sea. Ph.D. Thesis, Graduate University of Chinese Academy of Sciences, Shandong, China, 2008. (In Chinese). [Google Scholar]
- Jiang, H.; Cheng, H.Q.; Xu, H.G.; Arreguín, S.F.; Zetina, R.M.J.; Del, M.L.P.; Le, Q.W.J.F. Trophic controls of jellyfish blooms and links with fisheries in the East China Sea. Ecol. Model. 2008, 212, 492–503. [Google Scholar] [CrossRef]
- Yuan, X.C.; Yin, K.D.; Harrison, P.J. Bacterial production and respiration in subtropical Hong Kong waters: Influence of the Pearl River discharge and sewage effluent. Aquat. Microb. Ecol. 2010, 58, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Chao, S.Y. Circulation of the East China Sea, a numerical study. J. Oceanogr. 1990, 46, 273–295. [Google Scholar] [CrossRef]
- Li, M.; Xu, K.; Watanabe, M.; Chen, Z. Long-term variations in dissolved silicate, nitrogen, and phosphorus flux from the Yangtze River into the East China Sea and impacts on estuarine ecosystem. Estuar. Coast. Shelf Sci. 2007, 71, 3–12. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, J.N.; Zhang, Y.; Wang, A.P. Eukaryotic micro-plankton community diversity and characteristics of regional distribution in the Yellow Sea by ITS high-throughput sequencing. Environ. Scince 2018, 39, 2368–2379. [Google Scholar] [CrossRef]
- Condon, R.H.; Steinberg, D.K.; Del, P.A.; Bouvier, T.C.; Bronk, D.A.; Graham, W.M.; Ducklow, H.W. Jellyfish blooms result in a major microbial respiratory sink of carbon in marine systems. Proc. Natl. Acad. Sci. USA 2011, 108, 10225–10230. [Google Scholar] [CrossRef] [Green Version]
- Niggl, W.; Naumann, M.S.; Struck, U.; Manasrah, R.; Wild, C. Organic matter release by the benthic upside-down jellyfish Cassiopea sp. fuels pelagic food webs in coral reefs. J. Exp. Mar. Biol. Ecol. 2010, 384, 99–106. [Google Scholar] [CrossRef]
- Chen, X.C.; Huang, Y.; Mu, X.Y.; Zhu, L.Y.; Pu, Y.Q. Distribution characteristics of zooplankton in the Southern Yellow Sea in summer and winter. Period. Ocean Univ. China 2018, 48, 50–56. [Google Scholar]
- Gao, Q.; Xu, Z.; Zhuang, P. The relation between distribution of zooplankton and salinity in the Changjiang Estuary. Chin. J. Oceanol. Limnol. 2008, 26, 178–185. [Google Scholar] [CrossRef]
- Liang, D.; Uye, S. Population dynamics and production of the planktonic copepods in a eutrophic inlet of the Inland Sea of Japan. III. Paracalanus sp. Mar. Biol. 1996, 127, 219–227. [Google Scholar] [CrossRef]
- Wang, H.L.; Huang, B.Q.; Hong, H.S. Size-fractionated productivity and nutrient dynamics of phytoplankton in subtropical coastal environments. Hydrobiologia 1997, 352, 97–106. [Google Scholar] [CrossRef]
- Li, R.X.; Zhu, M.Y.; Wang, Z.L.; Shi, X.Y.; Chen, B.Z. Mesocosm experiment on competition between two HAB species in East China Sea. Chin. J. Appl. Ecol. 2003, 14, 1049–1054. [Google Scholar]
- Guo, S.; Feng, Y.; Wang, L.; Dai, M.; Liu, Z.; Bai, Y.; Sun, J. Seasonal variation in the phytoplankton community of a continental-shelf sea: The East China Sea. Mar. Ecol. Prog. 2014, 516, 103–126. [Google Scholar] [CrossRef] [Green Version]
- De Queiroz, A.R.; Flores Montes, M.; De Castro Melo, P.A.M.; Da Silva, R.A.; Koening, M.L. Vertical and horizontal distribution of phytoplankton around an oceanic archipelago of the Equatorial Atlantic. Mar. Biodivers. Rec. 2015, 8. [Google Scholar] [CrossRef]
- Wei, N.; Satheeswaran, T.; Jenkinson, I.R.; Xue, B.; Wei, Y.; Liu, H.; Sun, J. Factors driving the spatiotemporal variability in phytoplankton in the Northern South China Sea. Cont. Shelf Res. 2018, 162, 48–55. [Google Scholar] [CrossRef]
- Shiah, F.K.; Ducklow, H.W. Temperature regulation of heterotrophic bacterioplankton abundance, production, and specific growth rate in Chesapeake Bay. Limnol. Oceanogr. 1994, 39, 1243–1258. [Google Scholar] [CrossRef]
- Song, J.M. Biogeochemistry of China Marginal Seas; Shandong Science and Technology Press: Jinan, China, 2004. [Google Scholar]
- Pang, C.G.; Hu, D.X. Upwelling and sedimentation dynamics III: Coincidence of upwelling areas with mud patches in north hemisphere shelf seas. Chin. J. Oceanol. Limnol. 2002, 20, 101–106. [Google Scholar] [CrossRef]
- Tavernini, S.; Mura, G.; Rossetti, G. Factors Influencing the Seasonal Phenology and Composition of Zooplankton Communities in Mountain Temporary Pools. Int. Rev. Hydrobiol. 2005, 90, 358–375. [Google Scholar] [CrossRef]
- Le, F.; Sun, J.; Ning, X.; Song, S.; Cai, Y.; Liu, C. Phytoplankton in the northern south China sea in summer 2004. Oceanol. Limnol. Sin. 2006, 37, 238. [Google Scholar] [CrossRef]
- Conley, D.J.; Schelske, C.L.; Stoermer, E.F. Modification of the biogeochemical cycle of silica with eutrophication. Mar. Ecol. Prog. Ser. 1993, 101, 179–192. [Google Scholar] [CrossRef]
- Medinger, R.; Nolte, V.; Pandey, R.V.; Jost, S.; Ottenwalder, B.; Schlotterer, C.; Boenigk, J. Diversity in a hidden world: Potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 2010, 19, 32–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sime, N.T.; Grolière, C.A. Effets quantitatifs des fixateurs sur le stockage des ciliés d’eaux douces. Archiv. Protistenkd. 1991, 140, 109–120. [Google Scholar]








| Sample ID | OTUs | Chao | Shannon | Simpson | Coverage | |
|---|---|---|---|---|---|---|
| CWM–S | H_01S | 1392 | 1699 | 4.69 | 0.04 | 0.9963 |
| H_02S | 1159 | 1555 | 3.28 | 0.15 | 0.9957 | |
| H_03S | 1059 | 1370 | 3.40 | 0.12 | 0.9960 | |
| H_04S | 902 | 1204 | 3.06 | 0.19 | 0.9954 | |
| H_05S | 2260 | 2755 | 5.02 | 0.05 | 0.9935 | |
| Average | 1354 bc | 1717 a | 3.89 b | 0.11 a | 0.995 bc | |
| CWM–M | H_01M | 1548 | 1941 | 5.33 | 0.01 | 0.9943 |
| H_02M | 1536 | 1949 | 4.13 | 0.08 | 0.9953 | |
| H_03M | 1439 | 1833 | 4.21 | 0.09 | 0.9932 | |
| H_04M | 1360 | 1758 | 3.22 | 0.27 | 0.9944 | |
| H_05M | 2028 | 2512 | 5.76 | 0.01 | 0.9931 | |
| Average | 1582 ab | 1999 a | 4.53 ab | 0.09 a | 0994 c | |
| CWM–B | H_01B | 1680 | 1999 | 5.41 | 0.01 | 0.9955 |
| H_02B | 2050 | 2437 | 5.53 | 0.01 | 0.9938 | |
| H_03B | 2104 | 2425 | 5.45 | 0.02 | 0.9949 | |
| H_04B | 1887 | 2274 | 5.40 | 0.02 | 0.9924 | |
| H_05B | 2343 | 2897 | 5.19 | 0.04 | 0.9926 | |
| Average | 2013 a | 2406 a | 5.40 a | 0.02 a | 0994 c | |
| NW–S | H_06S | 1432 | 1687 | 5.07 | 0.02 | 0.9960 |
| H_07S | 330 | 379 | 3.82 | 0.06 | 0.9994 | |
| H_08S | 1442 | 1524 | 5.51 | 0.01 | 0.9986 | |
| H_09S | 350 | 419 | 3.03 | 0.24 | 0.9979 | |
| H_10S | 408 | 505 | 4.64 | 0.02 | 0.9970 | |
| Average | 792 cd | 903 b | 4.4 ab | 0.07 a | 0.998 a | |
| NW–M | H_06M | 1274 | 1508 | 4.64 | 0.05 | 0.9950 |
| H_07M | 335 | 380 | 4.13 | 0.04 | 0.9992 | |
| H_08M | 517 | 583 | 4.89 | 0.02 | 0.9989 | |
| H_09M | 536 | 677 | 4.49 | 0.03 | 0.9977 | |
| H_10M | 710 | 825 | 5.33 | 0.01 | 0.9965 | |
| Average | 674 d | 795 b | 4.70 ab | 0.03 a | 0.997 ab | |
| NW–B | H_06B | 1317 | 1636 | 4.65 | 0.05 | 0.9949 |
| H_07B | 488 | 591 | 4.54 | 0.02 | 0.9982 | |
| H_08B | 484 | 547 | 4.44 | 0.02 | 0.9991 | |
| H_09B | 704 | 915 | 5.27 | 0.01 | 0.9962 | |
| H_10B | 349 | 403 | 4.24 | 0.04 | 0.9988 | |
| Average | 668 d | 818 b | 4.63 ab | 0.03 a | 0.997 ab | |
| Environmental Parameters | CWM | NW | ||||
|---|---|---|---|---|---|---|
| CWM–S | CWM–M | CWM–B | NW–S | NW–M | NW–B | |
| Depth (m) | 4.92 ± 0.50 c | 35.82 ± 3.52 b | 75.16 ± 1.90 a | 4.42 ± 0.45 c | 17.13 ± 6.50 bc | 30.98 ± 10.72 b |
| pH | 8.09 ± 0.02 a | 7.99 ± 0.04 a | 7.78 ± 0.02 b | 8.07 ± 0.02 a | 8.02 ± 0.03 a | 8.00 ± 0.05 a |
| Salinity | 31.69 ± 0.12 abc | 32.30 ± 0.12 ab | 32.88 ± 0.48 a | 30.95 ± 0.34 c | 31.15 ± 0.46 bc | 31.24 ± 0.54 bc |
| Temperature | 21.24 ± 0.44 a | 15.72 ± 1.93 c | 9.55 ± 0.28 b | 22.13 ± 0.25 a | 20.62 ± 1.64 a | 20.05 ± 2.21 ab |
| Chl a (µg/L) | 0.64 ± 0.03 ab | 0.59 ± 0.14 ab | 0.21 ± 0.04 b | 1.18 ± 0.30 a | 1.16 ±0.33 a | 1.41 ± 0.33 a |
| NO2−-N (μmol/L) | 0.09 ± 0.02 a | 0.30 ± 0.09 a | 0.09 ± 0.01 a | 0.24 ± 0.07 a | 0.20 ± 0.06 a | 0.30 ± 0.16 a |
| NO3−-N (μmol/L) | 1.39 ± 0.40 b | 3.80 ± 0.66 b | 6.24 ± 0.32 ab | 9.10 ± 3.36 ab | 8.35 ± 2.53 ab | 13.63 ± 3.45 a |
| PO43−-P (μmol/L) | 0.09 ± 0.01 c | 0.32 ± 0.08 bc | 0.70 ± 0.05 a | 0.40 ± 0.11 b | 0.49 ± 0.08 ab | 0.50 ± 0.11 ab |
| SiO32−-Si (μmol/L) | 2.17 ± 0.26 c | 3.73 ± 0.32 bc | 8.95 ± 0.88 ab | 8.21 ± 2.60 ab | 9.05 ± 2.18 ab | 9.78 ± 1.82 a |
| NH4+-N (μmol/L) | 1.87 ± 0.34 a | 2.90 ± 0.50 a | 1.71 ± 0.35 a | 2.90 ± 0.60 a | 2.22 ± 0.57 a | 2.20 ± 0.44 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liu, Y.; Wu, C.; Fu, X.; Guo, C.; Li, L.; Sun, J. Characteristics of Eukaryotic Plankton Communities in the Cold Water Masses and Nearshore Waters of the South Yellow Sea. Diversity 2021, 13, 21. https://0-doi-org.brum.beds.ac.uk/10.3390/d13010021
Sun Y, Liu Y, Wu C, Fu X, Guo C, Li L, Sun J. Characteristics of Eukaryotic Plankton Communities in the Cold Water Masses and Nearshore Waters of the South Yellow Sea. Diversity. 2021; 13(1):21. https://0-doi-org.brum.beds.ac.uk/10.3390/d13010021
Chicago/Turabian StyleSun, Yanfeng, Yang Liu, Chao Wu, Xiaoting Fu, Congcong Guo, Liuyang Li, and Jun Sun. 2021. "Characteristics of Eukaryotic Plankton Communities in the Cold Water Masses and Nearshore Waters of the South Yellow Sea" Diversity 13, no. 1: 21. https://0-doi-org.brum.beds.ac.uk/10.3390/d13010021