Not the Cryptic Species: Diversity of Hipposideros gentilis (Chiroptera: Hipposideridae) in Indochina
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analyzed Specimens
2.2. Morphometric Analysis.
2.3. Molecular Analysis
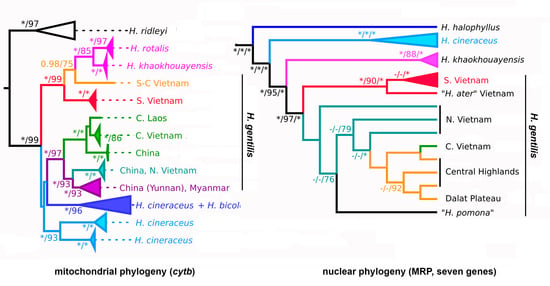
3. Results
3.1. Pattern of Variation in the mtDNA Marker
3.2. Pattern of Variation in nuDNA Markers
3.3. Patterns of Morphological Variation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. List of the GenBank Sequences of the Cytb Mitochondrial Gene, Used in the Analysis
Appendix B. List of the BOLD Process ID Numbers for Specimens Used in the Analysis
Appendix C. List of Specimens Used in the Morphometric Analysis. Specimens IDs Genotyped by at Least One Mitochondrial Gene Are in Italic, Specimens Genotyped by Nuclear Genes Are in Bold
References
- Foley, N.M.; Goodman, S.M.; Whelan, C.V.; Puechmaille, S.J.; Teeling, E. Towards navigating the Minotaur’s labyrinth: Cryptic diversity and taxonomic revision within the speciose genus Hipposideros (Hipposideridae). Acta Chiropterol. 2017, 19, 1–18. [Google Scholar] [CrossRef]
- Monadjem, A.; Soisook, P.; Thong, V.D.; Kingston, T. Family Hipposideridae (Old World Leaf-nosed Bats). In Handbook of the Mammals of the World; Wilson, D.E., Mittermeier, R.A., Eds.; Lynx Edictions: Barcelona, Spain, 2019; Volume 9, pp. 210–259. [Google Scholar]
- Douangboubpha, B.; Bumrungsri, S.; Soisook, P.; Murray, S.W.; Puechmaille, S.J.; Satasook, C.; Bu, S.S.H.; Harrison, D.L.; Bates, P.J.J. A taxonomic review of Hipposideros halophyllus, with additional information on H. ater and H. cineraceus (Chiroptera: Hipposideridae) from Thailand and Myanmar. Acta Chiropterol. 2010, 12, 29–50. [Google Scholar] [CrossRef]
- Douangboubpha, B.; Bumrungsri, S.; Soisook, P.; Satasook, C.; Thomas, N.M.; Bates, P.J.J. A taxonomic review of the Hipposideros bicolor species complex and H. pomona (Chiroptera: Hipposideridae) in Thailand. Acta Chiropterol. 2010, 12, 415–438. [Google Scholar] [CrossRef]
- Zhao, L.-Z.; Bu, Y.-Z.; Zhou, H.-X.; Zhou, H.-W.; Zhang, Z.-X.; Niu, H.-X. Differences in Hipposideros pomona from three geographical regions in China based on morphology and molecular sequences data. J. Mammal. 2015, 96, 1305–1316. [Google Scholar] [CrossRef] [Green Version]
- Corbet, G.B.; Hill, J.E. The Mammals of the Indomalayan Region: A Systematic Review; Oxford University Press: Oxford, UK, 1992; pp. 1–488. [Google Scholar]
- Francis, C.M. A Field Guide to the Mammals of South-East Asia; New Holland: London, UK, 2008; pp. 1–392. [Google Scholar]
- Kruskop, S.V. Bats of Vietnam. Checklist and an Identification Manual, 2nd ed.; KMK Ltd.: Moscow, Russia, 2013; pp. 1–300. [Google Scholar]
- Hill, J.E. A revision of the genus Hipposideros. Bull. Brit. Mus. Nat. Hist. Zool. 1963, 11, 1–129. [Google Scholar] [CrossRef]
- Hill, J.E.; Zubaid, A.; Davison, G.W.H. The taxonomy of leaf-nosed bats of the Hipposideros bicolor group (Chiroptera: Hipposideridae) from southeastern Asia. Mammalia 1986, 50, 536–540. [Google Scholar] [CrossRef]
- Dorst, J. Une nouvelle chauve-souris de l’Indochine française Paracoelops megalotis. Bull. Paris Mus. Nat. Hist., ser. 2 1947, 19, 436–437. [Google Scholar]
- Thong, V.D.; Dietz, C.; Denzinger, A.; Bates, P.J.J.; Puechmaille, S.J.; Callou, C.; Schnitzler, H.-U. Resolving a mammal mystery: The identity of Paracoelops megalotis (Chiroptera: Hipposideridae). Zootaxa 2012, 3505, 75–85. [Google Scholar] [CrossRef] [Green Version]
- Francis, C.M.; Borisenko, A.V.; Ivanova, N.V.; Eger, J.L.; Lim, B.K.; Guillén-Servent, A.; Kruskop, S.V.; Mackie, I.; Hebert, P.D.N. The Role of DNA Barcodes in Understanding and Conservation of Mammal Diversity in Southeast Asia. PLoS ONE 2010, 5, e12575. [Google Scholar] [CrossRef] [Green Version]
- Murray, S.W.; Campell, P.; Kingston, T.; Zubaid, A.; Francis, C.M.; Kunz, T.H. Molecular phylogeny of hipposiderid bats from Southeast Asia and evidence of cryptic diversity. Mol. Phylogenetics Evol. 2012, 62, 597–611. [Google Scholar] [CrossRef]
- Kruskop, S.V. The bacula of some bat species from Indo-China: Rhinolophoids (Chiroptera: Rhinolophidae, Hipposideridae). Plecotus et al. 2014, 17, 3–17. [Google Scholar]
- Topal, G. Bacula of some Old World leaf-nosed bats (Rhinolophidae and Hipposideridae, Chiroptera: Mammalia). Vertebr. Hung. 1975, 16, 21–53. [Google Scholar]
- Shrinivasulu, B.; Shrinivasulu, C. In plain sight: Bacular and noseleaf morphology supports distinct specific status of Roundleaf Bats Hipposideros pomona Andersen, 1918 and Hipposideros gentilis Andersen, 1918 (Chiroptera: Hipposideridae). J. Threat. Taxa 2018, 10, 12018–12026. [Google Scholar] [CrossRef]
- Srinivasulu, C.; Srinivasulu, B.; Srinivasulu, A. Hipposideros gentilis. The IUCN Red List of Threatened Species 2020. Available online: https://0-dx-doi-org.brum.beds.ac.uk/10.2305/IUCN.UK.2020-3.RLTS.T180991219A180991293.en (accessed on 25 February 2021).
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; pp. 1–1546. [Google Scholar]
- Matthee, C.A.; Burzlaff, J.D.; Taylor, J.F.; Davis, S.K. Mining the mammalian genome for artiodactyl systematics. Syst. Biol. 2001, 50, 1–24. [Google Scholar] [CrossRef]
- Artyushin, I.V.; Bannikova, A.A.; Lebedev, V.S.; Kruskop, S.V. Mitochondrial DNA relationships among North Palaearctic Eptesicus (Vespertilionidae, Chiroptera) and past hybridization between Common Serotine and Northern Bat. Zootaxa 2009, 2262, 40–52. [Google Scholar] [CrossRef]
- Igea, J.; Juste, J.; Castresana, J. Novel intron markers to study the phylogeny of closely related mammalian species. BMC Evol. Biol. 2010, 10, 369. [Google Scholar] [CrossRef] [Green Version]
- Yusefovich, A.P.; Artyushin, I.V.; Raspopova, A.A.; Bannikova, A.A.; Kruskop, S.V. An attempt to reconstruct the phylogeny of the Hipposideros leaf-nosed bats based on nuclear gene markers. Dokl. Biol. Sci. 2020, 493, 136–140. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Katoh, T.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Altekar, G.; Dwarkadas, S.; Huelsenbeck, J.P.; Ronquist, F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 2004, 20, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Baum, B.R. Combining trees as a way of combining datasets for phylogenetic inference, and the desirability of combining gene trees. Taxon 1992, 41, 3–10. [Google Scholar] [CrossRef]
- Ragan, M.A. Phylogenetic inference based on matrix representation of trees. Mol. Phyl. Evol. 1992, 1, 53–58. [Google Scholar] [CrossRef]
- Ragan, M.A. Matrix representation in reconstructing phylogenetic relationships among the eukaryotes. Bio Syst. 1992, 28, 47–55. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Bates, P.J.J.; Harrison, D.L. Bats of the Indian Subcontinent; Harrison Zoological Museum: Sevenoaks, UK, 1997; pp. 1–258. [Google Scholar]
- Andersen, K. Diagnoses of new bats of the families Rhinolophidae and Megadermatidae. Ann. Mag. Nat. Hist. 1918, 2, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Murray, S.W.; Khan, F.A.; Kingston, T.; Zubaid, A.; Campbell, P. A new species in the Hipposideros bicolor group (Chiroptera: Hipposideridae) from Peninsular Malaysia. Acta Chiropterol. 2018, 20, 1–29. [Google Scholar] [CrossRef]






| Mus. ID | Species | Locality | Accession | |
|---|---|---|---|---|
| S-190280 | A. stoliczkanus | N. Vietnam | Cat Ba | MZ219226 |
| S-167159 | H. cf. armiger | C. Vietnam | Quan Binh | MZ219210 |
| S-195483 | H. cf. griffini | S. Vietnam | Dong Nai | MZ219216 |
| S-186724 | H. cineraceus | S. Vietnam | Con Dao | MZ219220 |
| S-186725 | H. cineraceus | S. Vietnam | Con Dao | MZ219221 |
| S-186730 | H. cineraceus | S. Vietnam | Con Dao | MZ219222 |
| S-191867 | H. cineraceus | S. Vietnam | Dong Nai | MZ219218 |
| S-195484 | H. cineraceus | S. Vietnam | Dong Nai | MZ219219 |
| S-186567 | H. diadema | S. Vietnam | Binh Phuoc | MZ219211 |
| S-191868 | H. galeritus | S. Vietnam | Dong Nai | MZ219225 |
| S-167170 | H. gentilis | C. Vietnam | Quan Binh | MZ219215 |
| S-167171 | H. gentilis | C. Vietnam | Quan Binh | MZ219214 |
| S-167172 | H. gentilis | C. Vietnam | Quan Binh | MZ219223 |
| S-167173 | H. gentilis | C. Vietnam | Quan Binh | MZ219224 |
| S-190298 | H. gentilis | N. Vietnam | Cat Ba | MZ219212 |
| S-190301 | H. gentilis | N. Vietnam | Cat Ba | MZ219213 |
| S-190302 | H. gentilis | S. Vietnam | Bihn Chau | MZ219227 |
| S-191870 | H. gentilis | S. Vietnam | Dong Nai | MZ219217 |
| S-198154 | H. gentilis | SC. Vietnam | Gia Lai | MZ219228 |
| S-189221 | H. grandis | S. Vietnam | Dong Nai | MZ219209 |
| species | ID No | Reference | Locality | THY | ABHD | ROGDI | RAG2 | ACOX | COPS | SORBS |
|---|---|---|---|---|---|---|---|---|---|---|
| H. gentilis | S-167174 | original | C Vietnam | MZ219110 | MZ219140 | MZ219182 | ||||
| H. gentilis | S-190298 | original | N. Vietnam | MZ219096 | MZ219134 | |||||
| H. gentilis | S-190299 | original | N. Vietnam | MZ219097 | MZ219135 | |||||
| H. gentilis | S-190301 | original | N. Vietnam | MZ219118 | MZ219199 | MZ219133 | MZ219179 | |||
| H. gentilis | S-190302 | original | S Vietnam | MZ219098 | MZ219119 | MZ219200 | MZ219170 | MZ219137 | MZ219160 | MZ219180 |
| H. gentilis | S-191870 | original | S Vietnam | MZ219099 | MZ219122 | MZ219203 | MZ219172 | MZ219138 | MZ219159 | MZ219181 |
| H. gentilis | S-191871 | original | S Vietnam | MZ219123 | MZ219204 | MZ219139 | ||||
| H. gentilis | S-191909 | original | S Vietnam | MZ219124 | MZ219205 | MZ219173 | MZ219136 | |||
| H. gentilis | S-198253 | original | SC Vietnam | MZ219107 | MZ219175 | MZ219152 | MZ219191 | |||
| H. gentilis | S-198254 | original | SC Vietnam | MZ219108 | MZ219129 | MZ219207 | MZ219176 | MZ219153 | MZ219192 | |
| H. gentilis | S-198255 | original | SC Vietnam | MZ219109 | MZ219208 | MZ219177 | MZ219154 | MZ219193 | ||
| H. abae | ML162-210211- HIPABA | [1] | Mali | KP176357 | KP176214 | KP176320 | KP176020 | |||
| H. armiger | S-195483 | original | S Vietnam | MZ219094 | MZ219127 | MZ219206 | MZ219174 | MZ219132 | MZ219157 | MZ219190 |
| H. armiger | T-171109-1 | [1] | Vietnam | KP176354 | KP176210 | KP176317 | KP176016 | |||
| H. centralis | S-192897 | original | Ethiopia | MZ219105 | MZ219125 | MZ219149 | ||||
| H. cf. abae | S-189528 | original | Ethiopia | MZ219104 | MZ219116 | MZ219150 | ||||
| H. cf. grandis | S-195421 | original | SC Vietnam | MZ219093 | MZ219126 | MZ219131 | MZ219156 | MZ219189 | ||
| H. cf. ruber | ML29-310111 -HIPCAF | [1] | Mali | KP176358 | KP176215 | KP176321 | KP176021 | |||
| H. cineraceus | S-167179 | original | C Vietnam | MZ219111 | MZ219146 | |||||
| H. cineraceus | S-186724 | original | S Vietnam | MZ219100 | MZ219165 | MZ219143 | MZ219161 | MZ219185 | ||
| H. cineraceus | S-186725 | original | S Vietnam | MZ219101 | MZ219113 | MZ219166 | MZ219144 | MZ219186 | ||
| H. cineraceus | S-186730 | original | S Vietnam | MZ219102 | MZ219114 | MZ219195 | MZ219167 | MZ219145 | ||
| H. cineraceus | S-191867 | original | S Vietnam | MZ219103 | MZ219120 | MZ219201 | MZ219171 | MZ219147 | MZ219162 | MZ219183 |
| H. cineraceus | S-195484 | original | S Vietnam | MZ219128 | MZ219142 | MZ219184 | ||||
| H. diadema | 10.0011 | [1] | Thailand | KY552688 | KY552683 | KY552686 | ||||
| H. diadema | S-186567 | original | S. Vietnam | MZ219095 | MZ219112 | MZ219194 | MZ219164 | MZ219158 | ||
| H. galeritus | S-191868 | original | S. Vietnam | MZ219121 | MZ219202 | MZ219148 | MZ219187 | |||
| H. galeritus | T-090708-6 | [1] | Vietnam | KP176355 | KP176211 | KP176017 | ||||
| H. grandis | S-189221 | original | S Vietnam | MZ219092 | MZ219115 | MZ219196 | MZ219168 | MZ219130 | MZ219155 | MZ219178 |
| H. halophyllus | Hhal2 | [1] | Thailand | KP176359 | KP176216 | KP176322 | KP176022 | |||
| H. jonesi | ML155BIS-160211-HIPJON | [1] | Mali | KP176213 | KP176319 | KP176019 | ||||
| H. khaokhouayensis | S-190294 | original | N Vietnam | MZ219117 | MZ219198 | MZ219141 | MZ219163 | |||
| H. khaokhouayensis | T-070108-1 | [1] | Vietnam | KY552735 | KY552730 | KY552733 | ||||
| H. larvatus | Hlar29 | [1] | Thailand | KP176360 | KP176217 | KP176323 | KP176023 | |||
| H. “ater”* | T-250608-2 | [1] | Vietnam | KY552740 | KY552736 | |||||
| H. “pomona”* | T-180809-3 | [1] | Vietnam | KP176356 | KP176212 | KP176318 | KP176018 | |||
| A. stoliczkanus | S-190280 | original | N Vietnam | MZ219106 | MZ219197 | MZ219169 | MZ219151 | MZ219188 |
| H. rotalis | H. khaokhouayensis | H. cineraceus C. Vietnam | H. cineraceus S. Vietnam | H. cineraceus Malaysia | H. gentilis S. Vietnam | H. gentilis S-C. Vietnam | H. gentilis C. Vietnam | H. gentilis N. Vietnam | H. gentilis Yunnan | H. gentilis Hainan | H. gentilis Laos | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H. rotalis | 0.0054 | 0.0084 | 0.0093 | 0.0080 | 0.0072 | 0.0082 | 0.0086 | 0.0088 | 0.0082 | 0.0086 | 0.0085 | |
| H. kha | 0.0402 | 0.0087 | 0.0094 | 0.0081 | 0.0081 | 0.0087 | 0.0090 | 0.0088 | 0.0082 | 0.0091 | 0.0089 | |
| H. cin C. Vietnam | 0.1021 | 0.0968 | 0.0076 | 0.0071 | 0.0088 | 0.0093 | 0.0073 | 0.0083 | 0.0073 | 0.0079 | 0.0073 | |
| H. cin S. Vietnam | 0.0988 | 0.0977 | 0.0803 | 0.0076 | 0.0091 | 0.0094 | 0.0089 | 0.0087 | 0.0077 | 0.0086 | 0.0087 | |
| H. cin Malaysia | 0.1092 | 0.1068 | 0.0965 | 0.0989 | 0.0079 | 0.0087 | 0.0075 | 0.0079 | 0.0070 | 0.0080 | 0.0080 | |
| H. gen S. Vietnam | 0.0753 | 0.0771 | 0.0953 | 0.0991 | 0.1028 | 0.0095 | 0.0089 | 0.0088 | 0.0080 | 0.0089 | 0.0091 | |
| H. gen S-C. Vietnam | 0.0683 | 0.0726 | 0.0985 | 0.0964 | 0.1037 | 0.0874 | 0.0092 | 0.0093 | 0.0089 | 0.0092 | 0.0094 | |
| H. gen C. Vietnam | 0.1007 | 0.0960 | 0.0886 | 0.0933 | 0.0988 | 0.0957 | 0.0985 | 0.0076 | 0.0065 | 0.0056 | 0.0047 | |
| H. gen N. Vietnam | 0.0980 | 0.0961 | 0.0904 | 0.0944 | 0.1018 | 0.0921 | 0.0964 | 0.0739 | 0.0061 | 0.0082 | 0.0079 | |
| H. gen Yunnan | 0.0994 | 0.0949 | 0.0896 | 0.0874 | 0.0981 | 0.0933 | 0.0934 | 0.0663 | 0.0583 | 0.0069 | 0.0069 | |
| H. gen Hainan | 0.1039 | 0.0989 | 0.0891 | 0.0911 | 0.1033 | 0.0977 | 0.0951 | 0.0417 | 0.0781 | 0.0769 | 0.0058 | |
| H. gen Laos | 0.0976 | 0.0937 | 0.0862 | 0.0904 | 0.1040 | 0.0973 | 0.1020 | 0.0266 | 0.0797 | 0.0722 | 0.0412 |
| PC I | PC II | PC III | PC IV | |
|---|---|---|---|---|
| TL | 0.3874 | 0.5700 | 0.2348 | 0.5591 |
| CCL | 0.4756 | 0.6068 | 0.1781 | 0.4944 |
| MW | 0.0197 | 0.8554 | 0.0030 | 0.2787 |
| BCW | 0.1766 | 0.8479 | −0.2165 | −0.0034 |
| OH | −0.0287 | 0.3039 | 0.0392 | 0.6229 |
| ZW | 0.1389 | 0.3070 | −0.3332 | 0.6959 |
| POC | 0.1169 | 0.1589 | −0.7789 | 0.0429 |
| RW | 0.2931 | 0.2531 | 0.0622 | 0.6742 |
| RL | 0.4299 | 0.3268 | 0.5776 | −0.1496 |
| CC | 0.4059 | 0.0960 | −0.0271 | 0.7712 |
| MM | 0.3745 | 0.0619 | −0.4280 | 0.6985 |
| CM | 0.7334 | 0.3366 | 0.0206 | 0.4562 |
| PM | 0.7123 | 0.1227 | −0.1282 | 0.4650 |
| C | 0.8001 | −0.1206 | −0.0797 | 0.0967 |
| NO | −0.1033 | 0.5249 | 0.4612 | 0.2852 |
| CM | 0.6273 | 0.2283 | 0.0239 | 0.3402 |
| MDL | 0.4617 | 0.3784 | 0.0390 | 0.6815 |
| MDH | 0.1897 | −0.0661 | 0.0250 | 0.6867 |
| Eigenvalue | 8.0850 | 2.07005 | 1.4810 | 1.1372 |
| % Total | 44.9168 | 11.5003 | 8.2276 | 6.3176 |
| Cumulative variance | 44.9168 | 56.4171 | 64.6447 | 70.9622 |
| Training Sets | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1. Myanmar | 21.3396 | 28.9994 | 31.0698 | 19.3681 | 25.8214 | |
| 2. C Thailand | 0.0000 | 13.5448 | 15.6349 | 10.2886 | 15.3410 | |
| 3. NW Indochina | 0.0000 | 0.0000 | 3.1906 | 7.0932 | 34.5136 | |
| 4. NE Indochina/SE China | 0.0000 | 0.0000 | 0.4175 | 10.6141 | 40.3524 | |
| 5. C China | 0.0000 | 0.0002 | 0.0068 | 0.0000 | 20.9887 | |
| 6. S Indochina | 0.0000 | 0.0002 | 0.0000 | 0.0000 | 0,0000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuzefovich, A.P.; Artyushin, I.V.; Kruskop, S.V. Not the Cryptic Species: Diversity of Hipposideros gentilis (Chiroptera: Hipposideridae) in Indochina. Diversity 2021, 13, 218. https://0-doi-org.brum.beds.ac.uk/10.3390/d13050218
Yuzefovich AP, Artyushin IV, Kruskop SV. Not the Cryptic Species: Diversity of Hipposideros gentilis (Chiroptera: Hipposideridae) in Indochina. Diversity. 2021; 13(5):218. https://0-doi-org.brum.beds.ac.uk/10.3390/d13050218
Chicago/Turabian StyleYuzefovich, Alexander P., Ilya V. Artyushin, and Sergei V. Kruskop. 2021. "Not the Cryptic Species: Diversity of Hipposideros gentilis (Chiroptera: Hipposideridae) in Indochina" Diversity 13, no. 5: 218. https://0-doi-org.brum.beds.ac.uk/10.3390/d13050218