Brain-Targeted Delivery of Pre-miR-29b Using Lactoferrin-Stearic Acid-Modified-Chitosan/Polyethyleneimine Polyplexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Structural Characterization of CS/PEI-SA and CS/PEI-SA-Lf
2.2. Polyplexes Characterization
2.3. Polyplexes Cytotoxic Profile
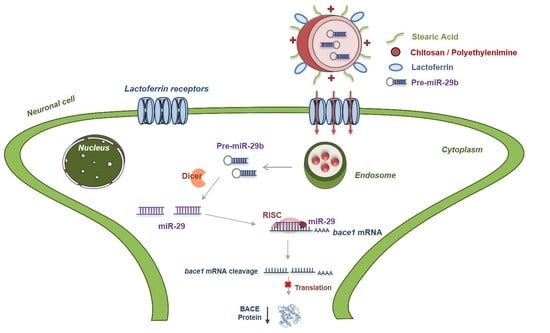
2.4. In Vitro Cellular Uptake and Intracellular Distribution of Pre-miR-29b-Loaded Complexes
2.5. Human BACE1 Gene Knockdown Using Pre-miR-29b-Loaded Complexes
2.6. Ability of Prepared Polyplexes to Cross in an In Vitro BBB Model
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Synthesis of CS and PEI Conjugated with SA
3.3. Functionalization of CS/PEI-SA with Lactoferrin
3.4. NMR Experiments
3.5. Preparation of Polymer-RNA Complexes (Polyplexes)
3.6. Gel Retardation Assay
3.7. Determination of the Encapsulation Efficiency
3.8. Scanning Electron Micrograph Morphology
3.9. Dynamic Light Scattering and Zeta Potential Analysis
3.10. Biological Activity
3.11. Cell Viability Assay
3.12. In Vitro Cellular Uptake—Cell Live Imaging
3.13. In Vitro Transfection Studies
3.14. Expression of Human BACE1 mRNA in N2a695 Cells by RT-qPCR
3.15. Transport Across an In Vitro BBB Model
3.15.1. BBB Transport Experiment
3.15.2. Immunofluorescence
3.16. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Isik, A.T. Late onset Alzheimer’s disease in older people. Clin. Interv. Aging 2010, 5, 307–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Alzheimer’s disease: Key developments support promising perspectives for therapy. Pharmacol. Res. 2019, 146, 104316. [Google Scholar] [CrossRef] [PubMed]
- Mullane, K.; Williams, M. Alzheimer’s therapeutics: Continued clinical failures question the validity of the amyloid hypothesis-but what lies beyond? Biochem. Pharmacol. 2013, 85, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Dorszewska, J.; Prendecki, M.; Oczkowska, A.; Dezor, M.; Kozubski, W. Molecular Basis of Familial and Sporadic Alzheimer’s Disease. Curr. Alzheimer Res. 2016, 13, 952–963. [Google Scholar] [CrossRef]
- Xheng, H.; Fridkin, M.; Youdim, M. From single to multitarged/network therapeutics in Alzheimer’s disease. Pharmaceuticals 2014, 7, 113–135. [Google Scholar]
- Ghiso, J.; Frangione, B. Amyloidosis and Alzheimer’s disease. Adv. Drug Deliv. Rev. 2002, 54, 1539–1551. [Google Scholar] [CrossRef]
- Provost, P. Interpretation and applicability of microRNA data to the context of Alzheimer’s and age-related diseases. Aging 2010, 2, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Schonrock, N.; Matamales, M.; Ittner, L.M.; Götz, J. MicroRNA networks surrounding APP and amyloid-β metabolism-implications for Alzheimer’s disease. Exp. Neurol. 2012, 235, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Vassar, R.; Kovacs, D.M.; Yan, R.; Wong, P.C. The beta-secretase enzyme BACE in health and Alzheimer’s disease: Regulation, cell biology, function, and therapeutic potential. J. Neurosci. 2009, 29, 12787–12794. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.L.; Vassar, R. The Alzheimer’s disease beta-secretase enzyme, BACE1. Mol. Neurodegener. 2007, 2, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, C.E.; Turner, A.J. Cell biology, regulation and inhibition of beta-secretase (BACE-1). FEBS J. 2009, 276, 1845–1859. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Basak, I.; Patil, K.S.; Alves, G.; Larsen, J.P.; Møller, S.G. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci. 2016, 73, 811–827. [Google Scholar] [CrossRef]
- Abe, M.; Bonini, N.M. MicroRNAs and neurodegeneration: Role and impact. Trends Cell Biol. 2013, 23, 30–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodall, E.F.; Heath, P.R.; Bandmann, O.; Kirby, J.; Shaw, P.J. Neuronal dark matter: The emerging role of microRNAs in neurodegeneration. Front. Cell. Neurosci. 2013, 7, 178. [Google Scholar] [CrossRef] [Green Version]
- Dunkel, P.; Chai, C.L.; Sperlágh, B.; Huleatt, P.B.; Mátyus, P. Clinical utility of neuroprotective agents in neurodegenerative diseases: Current status of drug development for Alzheimer’s, Parkinson’s and Huntington’s diseases, and amyotrophic lateral sclerosis. Expert Opin. Investig. Drugs 2012, 21, 1267–1308. [Google Scholar] [CrossRef]
- Pereira, P.; Queiroz, J.A.; Figueiras, A.; Sousa, F. Current progress on microRNAs-based therapeutics in neurodegenerative diseases. Wiley Interdiscip. Rev. RNA 2017, 8, e1409. [Google Scholar] [CrossRef]
- Pereira, P.A.; Tomás, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant pre-miR-29b for Alzheimer’s disease therapeutics. Sci. Rep. 2016, 6, 19946. [Google Scholar] [CrossRef]
- Pardridge, W.M. CSF, blood-brain barrier, and brain drug delivery. Expert Opin. Drug Deliv. 2016, 13, 963–975. [Google Scholar] [CrossRef]
- Dong, X. Current Strategies for Brain Drug Delivery. Theranostics 2018, 8, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sharma, A.R.; Lee, S.S.; Bhattacharya, M.; Nam, J.S.; Chakraborty, C. Advances in nanocarriers enabled brain targeted drug delivery across blood brain barrier. Int. J. Pharm. 2019, 559, 360–372. [Google Scholar] [CrossRef]
- Pardridge, W.M. Delivery of Biologics Across the Blood-Brain Barrier with Molecular Trojan Horse Technology. BioDrugs 2017, 31, 503–519. [Google Scholar] [CrossRef]
- Löscher, W.; Potschka, H. Role of drug efflux transporters in the brain for drug disposition and treatment of brain diseases. Prog. Neurobiol. 2005, 76, 22–76. [Google Scholar] [CrossRef] [PubMed]
- Descamps, L.; Dehouck, M.P.; Torpier, G.; Cecchelli, R. Receptor-mediated transcytosis of transferrin through blood-brain barrier endothelial cells. Am. J. Physiol. 1996, 270, H1149–H1158. [Google Scholar]
- Visser, C.C.; Stevanović, S.; Heleen Voorwinden, L.; Gaillard, P.J.; Crommelin, D.J.; Danhof, M.; De Boer, A.G. Validation of the transferrin receptor for drug targeting to brain capillary endothelial cells in vitro. J. Drug Target. 2004, 12, 145–150. [Google Scholar] [CrossRef]
- Fillebeen, C.; Descamps, L.; Dehouck, M.P.; Fenart, L.; Benaïssa, M.; Spik, G.; Cecchelli, R.; Pierce, A. Receptor-mediated transcytosis of lactoferrin through the blood-brain barrier. J. Biol. Chem. 1999, 274, 7011–7017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candela, P.; Gosselet, F.; Miller, F.; Buee-Scherrer, V.; Torpier, G.; Cecchelli, R.; Fenart, L. Physiological pathway for low-density lipoproteins across the blood-brain barrier: Transcytosis through brain capillary endothelial cells in vitro. Endothelium 2008, 15, 254–264. [Google Scholar] [CrossRef]
- Dehouck, B.; Fenart, L.; Dehouck, M.P.; Pierce, A.; Torpier, G.; Cecchelli, R. A new function for the LDL receptor: Transcytosis of LDL across the blood-brain barrier. J. Cell Biol. 1997, 138, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Huwyler, J.; Wu, D.; Pardridge, W.M. Brain drug delivery of small molecules using immunoliposomes. Proc. Natl. Acad. Sci. USA 1996, 93, 14164–14169. [Google Scholar] [CrossRef] [Green Version]
- Swami, A.; Goyal, R.; Tripathi, S.K.; Singh, N.; Katiyar, N.; Mishra, A.K.; Gupta, K.C. Effect of homobifunctional crosslinkers on nucleic acids delivery ability of PEI nanoparticles. Int. J. Pharm. 2009, 374, 125–138. [Google Scholar] [CrossRef]
- Sarvaiya, J.; Agrawal, Y.K. Chitosan as a suitable nanocarrier material for anti-Alzheimer drug delivery. Int. J. Biol. Macromol. 2015, 72, 454–465. [Google Scholar] [CrossRef] [PubMed]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.P.; Paz, E.; Conneely, O.M. Multifunctional roles of lactoferrin: A critical overview. Cell. Mol. Life. Sci. 2005, 62, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Q.; Ke, W.L.; Qu, Y.H.; Zhu, J.H.; Pei, Y.Y.; Jiang, C. Characterization of lactoferrin receptor in brain endothelial capillary cells and mouse brain. J. Biomed. Sci. 2007, 14, 121–128. [Google Scholar] [CrossRef]
- Ji, B.; Maeda, J.; Higuchi, M.; Inoue, K.; Akita, H.; Harashima, H.; Suhara, T. Pharmacokinetics and brain uptake of lactoferrin in rats. Life Sci. 2006, 78, 851–855. [Google Scholar] [CrossRef]
- Van de Looij, Y.; Ginet, V.; Chatagner, A.; Toulotte, A.; Somm, E.; Hüppi, P.S.; Sizonenko, S.V. Lactoferrin during lactation protects the immature hypoxic-ischemic rat brain. Ann. Clin. Transl. Neurol. 2014, 1, 955–967. [Google Scholar] [CrossRef]
- Rousseau, E.; Michel, P.P.; Hirsch, E.C. The iron-binding protein lactoferrin protects vulnerable dopamine neurons from degeneration by preserving mitochondrial calcium homeostasis. Mol. Pharmacol. 2013, 84, 888–898. [Google Scholar] [CrossRef] [Green Version]
- Iwamaru, Y.; Shimizu, Y.; Imamura, M.; Murayama, Y.; Endo, R.; Tagawa, Y.; Ushiki-Kaku, Y.; Takenouchi, T.; Kitani, H.; Mohri, S.; et al. Lactoferrin induces cell surface retention of prion protein and inhibits prion accumulation. J. Neurochem. 2008, 107, 636–646. [Google Scholar] [CrossRef]
- Guo, C.; Yang, Z.H.; Zhang, S.; Chai, R.; Xue, H.; Zhang, Y.H.; Li, J.Y.; Wang, Z.Y. Intranasal Lactoferrin Enhances α-Secretase-Dependent Amyloid Precursor Protein Processing via the ERK1/2-CREB and HIF-1α Pathways in an Alzheimer’s Disease Mouse Model. Neuropsychopharmacology 2017, 42, 2504–2515. [Google Scholar] [CrossRef] [Green Version]
- Onishi, H.; Koyama, K.; Sakata, O.; Machida, Y. Preparation of chitosan/alginate/calcium complex microparticles loaded with lactoferrin and their efficacy on carrageenan-induced edema in rats. Drug Dev. Ind. Pharm. 2010, 36, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Ke, W.; Han, L.; Liu, Y.; Shao, K.; Jiang, C.; Pei, Y. Lactoferrin-modified nanoparticles could mediate efficient gene delivery to the brain in vivo. Brain Res. Bull. 2010, 81, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Wang, T.; Xu, Q.; Chang, Y.; Tu, H.; Zheng, Y.; Zhang, J.; Xu, Y.; Yang, J.; Yuan, H.; et al. Genotoxicity evaluation of stearic acid grafted chitosan oligosaccharide nanomicelles. Mutat. Res. Toxicol. Environ. Mutagen. 2013, 751, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.T.; Du, Y.Z.; Yuan, H.; Hu, F.Q. Brain-targeting study of stearic acid-grafted chitosan micelle drug-delivery system. Int. J. Nanomed. 2012, 7, 3235–3244. [Google Scholar]
- Chuanxu, Y.; Shan, G.; Jørgen, K. Folic acid conjugated chitosan for targeted delivery of siRNA to activated macrophages in vitro and in vivo. J. Mater. Chem. B 2014, 2, 8608–8615. [Google Scholar]
- Yuan, H.; Lu, L.J.; Du, Y.Z.; Hu, F.Q. Stearic acid-g-chitosan polymeric micelle for oral drug delivery: In vitro transport and in vivo absorption. Mol. Pharm. 2011, 8, 225–238. [Google Scholar] [CrossRef]
- Jingou, J.; Danjun, W.; Li, L.; Chen, J.; Xiu, Y. Preparation, evaluation, and in vitro release of folic acid conjugated O-carboxymethyl chitosan nanoparticles loaded with methotrexate. J. Appl. Polym. Sci. 2011, 125, e208–e215. [Google Scholar]
- Pereira, P.; Jorge, A.F.; Martins, R.; Pais, A.A.; Sousa, F.; Figueiras, A. Characterization of polyplexes involving small RNA. J. Colloid Interface Sci. 2012, 387, 84–94. [Google Scholar] [CrossRef]
- Huang, R.; Ke, W.; Han, L.; Liu, Y.; Shao, K.; Ye, L.; Lou, J.; Jiang, C.; Pei, Y. Brain-targeting mechanisms of lactoferrin-modified DNA-loaded nanoparticles. J. Cereb. Blood Flow Metab. 2009, 29, 1914–1923. [Google Scholar] [CrossRef] [Green Version]
- Meng, Q.; Wang, A.; Hua, H.; Jiang, Y.; Wang, Y.; Mu, H.; Wu, Z.; Sun, K. Intranasal delivery of Huperzine A to the brain using lactoferrin-conjugated N-trimethylated chitosan surface-modified PLGA nanoparticles for treatment of Alzheimer’s disease. Int. J. Nanomed. 2018, 13, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhou, K.; Wang, R.; Liu, Y.; Kwak, Y.D.; Ma, T.; Thompson, R.C.; Zhao, Y.; Smith, L.; Gasparini, L.; et al. Antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer’s amyloid peptides via up-regulating BACE1 transcription. Proc. Natl. Acad. Sci. USA 2009, 106, 3907–3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, I.J.; Iyer, M.; Wagner, E.; Gambhir, S.S. Optical imaging of transferrin targeted PEI/DNA complexes in living subjects. Gene Ther. 2003, 10, 758–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Müller, K.; Kessel, E.; Reinhard, S.; He, D.; Klein, P.M.; Höhn, M.; Rödl, W.; Kempter, S.; Wagner, E. Targeted siRNA Delivery Using a Lipo-Oligoaminoamide Nanocore with an Influenza Peptide and Transferrin Shell. Adv. Healthc. Mater. 2016, 5, 1493–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huwyler, J.; Froidevaux, S.; Roux, F.; Eberle, A.N. Characterization of transferrin receptor in an immortalized cell line of rat brain endothelial cells, RBE4. J. Recept. Signal Transduct. 1999, 19, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, A.; Skjørringe, T.; Johnsen, K.B.; Siupka, P.; Thomsen, L.B.; Nielsen, M.S.; Thomsen, L.L.; Moos, T. Expression of Iron-Related Proteins at the Neurovascular Unit Supports Reduction and Reoxidation of Iron for Transport Through the Blood-Brain Barrier. Mol. Neurobiol. 2016, 53, 7237–7253. [Google Scholar] [CrossRef]
- Roux, F.; Couraud, P.O. Rat brain endothelial cell lines for the study of blood-brain barrier permeability and transport functions. Cell. Mol. Neurobiol. 2005, 25, 41–58. [Google Scholar] [CrossRef] [Green Version]
- Reis, K.; Hälldin, J.; Fernaeus, S.; Pettersson, C.; Land, T. NADPH oxidase inhibitor diphenyliodonium abolishes lipopolysaccharide-induced down-regulation of transferrin receptor expression in N2a and BV-2 cells. J. Neurosci. Res. 2006, 84, 1047–1052. [Google Scholar] [CrossRef]
- Balbuena, P.; Li, W.; Ehrich, M. Assessments of tight junction proteins occludin, claudin 5 and scaffold proteins ZO1 and ZO2 in endothelial cells of the rat blood-brain barrier: Cellular responses to neurotoxicants malathion and lead acetate. Neurotoxicology 2011, 32, 58–67. [Google Scholar] [CrossRef]
- Huang, R.; Ke, W.; Liu, Y.; Jiang, C.; Pei, Y. The use of lactoferrin as a ligand for targeting the polyamidoamine-based gene delivery system to the brain. Biomaterials 2008, 29, 238–246. [Google Scholar] [CrossRef]
- Song, Y.; Du, D.; Li, L.; Xu, J.; Dutta, P.; Lin, Y. In Vitro Study of Receptor-Mediated Silica Nanoparticles Delivery across Blood-Brain Barrier. ACS Appl. Mater. Interfaces 2017, 9, 20410–20416. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.Y.; Chen, W.J.; Lee, W.Y.; Lo, S.T.; Lee, T.W.; Lo, J.M. In vitro and in vivo evaluation of lactoferrin-conjugated liposomes as a novel carrier to improve the brain delivery. Int. J. Mol. Sci. 2013, 14, 2862–2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, Y.C.; Cheng, S.J. Brain targeted delivery of carmustine using solid lipid nanoparticles modified with tamoxifen and lactoferrin for antitumor proliferation. Int. J. Pharm. 2016, 499, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Ajazuddin; Tripathi, D.K.; Saraf, S.; Saraf, S.; Antimisiaris, S.G.; Mourtas, S.; Hammarlund-Udenaes, M.; Alexander, A. Recent advancements in liposomes targeting strategies to cross blood-brain barrier (BBB) for the treatment of Alzheimer’s disease. J. Control. Release 2017, 260, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Pedro, A.Q.; Tomás, J.; Maia, C.J.; Queiroz, J.A.; Figureiras, R.; Sousa, F. Advances in time course extracellular production of human pre-miR-29b from Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 2016, 100, 3723–3734. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Sousa, Â.; Queiroz, J.; Correia, I.; Figueiras, A.; Sousa, F. Purification of pre-miR-29 by arginine-affinity chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014, 951–952, 16–23. [Google Scholar] [CrossRef]
- Re, F.; Cambianica, I.; Zona, C.; Sesana, S.; Gregori, M.; Rigolio, R.; La Ferla, B.; Nicotra, F.; Forloni, G.; Cagnotto, A.; et al. Functionalization of liposomes with ApoE-derived peptides at different density affects cellular uptake and drug transport across a blood-brain barrier model. Nanomedicine 2011, 7, 551–559. [Google Scholar] [CrossRef]








| Polyplexes | Charge Ratio | Z-Average Diameter (nm) | Zeta Potential (mV) | EE (%) |
|---|---|---|---|---|
| CS-SA-Lf/pre-miR-29b | 35 | 325.60 ± 30.99 | +33.50 ± 2.31 | 96.35 ± 3.56 |
| 40 | 432.50 ± 50.72 | +37.26 ± 2.49 | 92.36 ± 6.70 | |
| PEI-SA-Lf/pre-miR-29b | 15 | 289.40 ± 39.12 | +10.74 ± 1.07 | 96.06 ± 2.81 |
| 20 | 463.45 ± 47.68 | +21.01 ± 3.12 | 94.44 ± 7.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, P.; Barreira, M.; Cruz, C.; Tomás, J.; Luís, Â.; Pedro, A.Q.; Queiroz, J.A.; Sousa, F. Brain-Targeted Delivery of Pre-miR-29b Using Lactoferrin-Stearic Acid-Modified-Chitosan/Polyethyleneimine Polyplexes. Pharmaceuticals 2020, 13, 314. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13100314
Pereira P, Barreira M, Cruz C, Tomás J, Luís Â, Pedro AQ, Queiroz JA, Sousa F. Brain-Targeted Delivery of Pre-miR-29b Using Lactoferrin-Stearic Acid-Modified-Chitosan/Polyethyleneimine Polyplexes. Pharmaceuticals. 2020; 13(10):314. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13100314
Chicago/Turabian StylePereira, Patrícia, Maria Barreira, Carla Cruz, Joana Tomás, Ângelo Luís, Augusto Q. Pedro, João A. Queiroz, and Fani Sousa. 2020. "Brain-Targeted Delivery of Pre-miR-29b Using Lactoferrin-Stearic Acid-Modified-Chitosan/Polyethyleneimine Polyplexes" Pharmaceuticals 13, no. 10: 314. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13100314