14-3-3σ and Its Modulators in Cancer
Abstract
:1. Introduction
2. 14-3-3σ (Stratifin, or Sfn)
3. Role of 14-3-3σ in Cancer
4. 14-3-3σ PPI Modulators
4.1. 14-3-3σ PPI Stabilizers
4.2. 14-3-3σ PPI Inhibitors
4.2.1. Phosphonate- and Phosphate-Type Inhibitors of 14-3-3σ
4.2.2. Non-Phosphonate-Type Inhibitors of 14-3-3σ
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ballone, A.; Centorrino, F.; Ottmann, C. 14-3-3: A case study in PPI modulation. Molecules 2018, 23, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Cui, L.; Zeng, Y.; Song, W.; Gaur, U.; Yang, M. 14-3-3 proteins are on the crossroads of cancer, aging, and age-related neurodegenerative disease. Int. J. Mol. Sci. 2019, 20, 3518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugaeva, K.V.; Titterington, J.; Sotnikov, D.V.; Maksimov, E.G.; Antson, A.A.; Sluchanko, N.N. Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family. bioRxiv 2020, 287, 3944–3966. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.W. Specific acidic proteins of the nervous system. In Physiological and Biochemical Aspects of Nervous Integration; Prentice-Hall: New York, NY, USA, 1967; pp. 343–359. [Google Scholar]
- Aitken, A.; Collinge, D.; Van Heusden, B.; Isobe, T.; Roseboom, P.; Rosenfeld, G.; Soll, J. 14-3-3 proteins. a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 1992, 17, 498–501. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ren, Y.; Li, J.; Ji, Z.; Chen, F.; Wang, X. Identification of the 14-3-3 β/α-A protein as a novel maternal peptidoglycan-binding protein that protects embryos of zebrafish against bacterial infections. Dev. Comp. Immunol. 2020, 114, 103867. [Google Scholar] [CrossRef] [PubMed]
- De, S. The 14-3-3 (YWHA) Proteins in mammalian reproduction. Int. Ann. Sci. 2020, 10, 52–59. [Google Scholar] [CrossRef]
- Nathan, K.G.; Lal, S.K. The multifarious role of 14-3-3 family of proteins in viral replication. Viruses 2020, 12, 436. [Google Scholar] [CrossRef]
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. [Google Scholar] [CrossRef]
- Pennington, K.L.; Chan, T.Y.; Torres, M.P.; Andersen, J.L. The dynamic and stress-adaptive signaling hub of 14-3-3: Emerging mechanisms of regulation and context-dependent protein-protein interactions. Oncogene 2018, 37, 5587–5604. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Cuevas, E.; Raymick, J.; Kanungo, J.; Sarkar, S. Downregulation of 14-3-3 proteins in Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 32–40. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Umahara, T.; Hirokawa, K.; Hanyu, H.; Uchihara, T. 14-3-3 protein sigma isoform co-localizes with phosphorylated α-synuclein in Lewy bodies and Lewy neurites in patients with Lewy body disease. Neurosci. Lett. 2018, 674, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shakes, D.C. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 1996, 43, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001, 127, 142–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Heusden, G.P.H. 14-3-3 proteins: Insights from genome-wide studies in yeast. Genomics 2009, 94, 287–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Heusden, G.P.; Steensma, H.Y. Yeast 14-3-3 proteins. Yeast 2006, 23, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.H.; Ley, S.; Aitken, A. Isoforms of 14-3-3 protein can form homo-and heterodimers in vivo and in vitro: Implications for function as adapter proteins. FEBS Lett. 1995, 368, 55–58. [Google Scholar] [CrossRef] [Green Version]
- Aitken, A.; Baxter, H.; Dubois, T.; Clokie, S.; Mackie, S.; Mitchell, K.; Peden, A.; Zemlickova, E. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 2002, 30, 351–360. [Google Scholar] [CrossRef]
- Gardino, A.K.; Smerdon, S.J.; Yaffe, M.B. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: A comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin. Cancer Biol. 2006, 16, 173–182. [Google Scholar] [CrossRef]
- Chen, Y.; Ruggeri, Z.M.; Du, X. 14-3-3 proteins in platelet biology and glycoprotein Ib-IX signaling. Blood 2018, 131, 2436–2448. [Google Scholar] [CrossRef] [Green Version]
- Lalle, M.; Fiorillo, A. The protein 14-3-3: A functionally versatile molecule in Giardia duodenalis. In Advances in Parasitology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 106, pp. 51–103. [Google Scholar]
- Cotelle, V.; Leonhardt, N. 14-3-3 proteins in guard cell signaling. Front. Plant Sci. 2016, 6, 1210. [Google Scholar] [CrossRef] [Green Version]
- Ormancey, M.; Thuleau, P.; Mazars, C.; Cotelle, V. CDPKs and 14-3-3 proteins: Emerging duo in signaling. Trends Plant Sci. 2017, 22, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Rittinger, K.; Budman, J.; Xu, J.; Volinia, S.; Cantley, L.C.; Smerdon, S.J.; Gamblin, S.J.; Yaffe, M.B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 1999, 4, 153–166. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Gusev, N.B. Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J. 2017, 284, 1279–1295. [Google Scholar] [CrossRef] [Green Version]
- Tugaeva, K.V.; Kalacheva, D.I.; Cooley, R.B.; Strelkov, S.V.; Sluchanko, N.N. Concatenation of 14-3-3 with partner phosphoproteins as a tool to study their interaction. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, M.B.; Rittinger, K.; Volinia, S.; Caron, P.R.; Aitken, A.; Leffers, H.; Gamblin, S.J.; Smerdon, S.J.; Cantley, L.C. The structural basis for 14-3-3: Phosphopeptide binding specificity. Cell 1997, 91, 961–971. [Google Scholar] [CrossRef] [Green Version]
- Coblitz, B.; Shikano, S.; Wu, M.; Gabelli, S.B.; Cockrell, L.M.; Spieker, M.; Hanyu, Y.; Fu, H.; Amzel, L.M.; Li, M. C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J. Biol. Chem. 2005, 280, 36263–36272. [Google Scholar] [CrossRef] [Green Version]
- Trcka, F.; Durech, M.; Vankova, P.; Vandova, V.; Simoncik, O.; Kavan, D.; Vojtesek, B.; Muller, P.; Man, P. The interaction of the mitochondrial protein importer TOMM34 with HSP70 is regulated by TOMM34 phosphorylation and binding to 14-3-3 adaptors. J. Biol. Chem. 2020, 295, 8928–8944. [Google Scholar] [CrossRef]
- Petosa, C.; Masters, S.C.; Bankston, L.A.; Pohl, J.; Wang, B.; Fu, H.; Liddington, R.C. 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 1998, 273, 16305–16310. [Google Scholar] [CrossRef] [Green Version]
- Masters, S.C.; Pederson, K.J.; Zhang, L.; Barbieri, J.T.; Fu, H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 1999, 38, 5216–5221. [Google Scholar] [CrossRef]
- Henriksson, M.L.; Trollér, U.; Hallberg, B. 14-3-3 proteins are required for the inhibition of Ras by exoenzyme S. Biochem. J. 2000, 349, 697–701. [Google Scholar] [CrossRef] [Green Version]
- Mils, V.; Baldin, V.; Goubin, F.; Pinta, I.; Papin, C.; Waye, M.; Eychene, A.; Ducommun, B. Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene 2000, 19, 1257–1265. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Lin, H.; Shamim, M.; Schlaepfer, W.W.; Cañete-Soler, R. Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 2001, 276, 41318–41324. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Ren, J.; He, X.; Chen, H.; Wei, T.; Feng, W. YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization. Autophagy 2019, 15, 1017–1030. [Google Scholar] [CrossRef]
- Bonnefoy-Bérard, N.; Liu, Y.C.; von Willebrand, M.; Sung, A.; Elly, C.; Mustelin, T.; Yoshida, H.; Ishizaka, K.; Altman, A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc. Natl. Acad. Sci. USA 1995, 92, 10142–10146. [Google Scholar] [CrossRef] [Green Version]
- Garnett, M.J.; Rana, S.; Paterson, H.; Barford, D.; Marais, R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 2005, 20, 963–969. [Google Scholar] [CrossRef]
- Rushworth, L.K.; Hindley, A.D.; O’Neill, E.; Kolch, W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol. Cell. Biol. 2006, 26, 2262–2272. [Google Scholar] [CrossRef] [Green Version]
- Ford, J.C.; Al-Khodairy, F.; Fotou, E.; Sheldrick, K.S.; Griffiths, D.; Carr, A.M. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 1994, 265, 533–535. [Google Scholar] [CrossRef]
- Peng, C.-Y.; Graves, P.R.; Thoma, R.S.; Wu, Z.; Shaw, A.S.; Piwnica-Worms, H. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997, 277, 1501–1505. [Google Scholar] [CrossRef]
- Sanchez, Y.; Wong, C.; Thoma, R.S.; Richman, R.; Wu, Z.; Piwnica-Worms, H.; Elledge, S.J. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science 1997, 277, 1497–1501. [Google Scholar] [CrossRef]
- Bulavin, D.V.; Higashimoto, Y.; Demidenko, Z.N.; Meek, S.; Graves, P.; Phillips, C.; Zhao, H.; Moody, S.A.; Appella, E.; Piwnica-Worms, H. Dual phosphorylation controls Cdc25 phosphatases and mitotic entry. Nat. Cell Biol. 2003, 5, 545–551. [Google Scholar] [CrossRef]
- Chen, M.-S.; Ryan, C.E.; Piwnica-Worms, H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 2003, 23, 7488–7497. [Google Scholar] [CrossRef] [Green Version]
- Samuel, T.; Weber, H.O.; Rauch, P.; Verdoodt, B.; Eppel, J.-T.; McShea, A.; Hermeking, H.; Funk, J.O. The G2/M regulator 14-3-3ς prevents apoptosis through sequestration of Bax. J. Biol. Chem. 2001, 276, 45201–45206. [Google Scholar] [CrossRef] [Green Version]
- Sunayama, J.; Tsuruta, F.; Masuyama, N.; Gotoh, Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005, 170, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Zha, J.; Harada, H.; Yang, E.; Jockel, J.; Korsmeyer, S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell 1996, 87, 619–628. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, J.; Fu, H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 8511–8515. [Google Scholar] [CrossRef] [Green Version]
- Prasad, G.L.; Valverius, E.M.; McDuffie, E.; Cooper, H.L. Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 1992, 3, 507–513. [Google Scholar]
- Leffers, H.; Madsen, P.; Rasmussen, H.H.; Honore, B.; Andersen, A.H.; Walbum, E.; Vandekerckhove, J.; Celis, J.E. Molecular cloning and expression of the transformation sensitive epithelial marker stratifin: A member of a protein family that has been involved in the protein kinase C signalling pathway. J. Mol. Biol. 1993, 231, 982–998. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, M.; Huang, W. 14-3-3 σ: A potential biomolecule for cancer therapy. Clin. Chim. Acta 2020, 511, 50–58. [Google Scholar] [CrossRef]
- Suárez-Bonnet, A.; Priestnall, S.; Ramírez, G.; Molín, J.; Jaber, J. Aberrant expression of cell cycle regulator 14-3-3-σ and E-cadherin in a metastatic cholangiocarcinoma in a vervet monkey (Chlorocebus pygerythrus). J. Comp. Pathol. 2020, 179, 25–30. [Google Scholar] [CrossRef]
- Benzinger, A.; Muster, N.; Koch, H.B.; Yates, J.R.; Hermeking, H. Targeted proteomic analysis of 14-3-3ς, a p53 effector commonly silenced in cancer. Mol. Cell. Proteom. 2005, 4, 785–795. [Google Scholar] [CrossRef] [Green Version]
- Benzinger, A.; Popowicz, G.M.; Joma, K.J.; Majumdar, S.; Holak, T.A.; Hermeking, H. The crystal structure of the non-liganded 14-3-3σ protein: Insights into determinants of isoform specific ligand binding and dimerization. Cell Res. 2005, 15, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilker, E.W.; Grant, R.A.; Artim, S.C.; Yaffe, M.B. A structural basis for 14-3-3σ functional specificity. J. Biol. Chem. 2005, 280, 18891–18898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.-D.; Huang, Y.-F.; Pan, Y.-J.; Huang, L.-K.; Liao, Y.-Y.; Lin, W.-H.; Liu, T.-Y.; Lee, C.-H.; Pan, R.-L. Regulation of H+-pyrophosphatase by 14-3-3 proteins from Arabidopsis thaliana. J. Membr. Biol. 2018, 251, 263–276. [Google Scholar] [CrossRef] [PubMed]
- McFerrin, M.B.; Chi, X.; Cutter, G.; Yacoubian, T.A. Dysregulation of 14-3-3 proteins in neurodegenerative diseases with Lewy body or Alzheimer pathology. Ann. Clin. Transl. Neurol. 2017, 4, 466–477. [Google Scholar] [CrossRef]
- Hu, G.; Li, H.; Liu, J.-Y.; Wang, J. Insight into conformational change for 14-3-3σ protein by molecular dynamics simulation. Int. J. Mol. Sci. 2014, 15, 2794–2810. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Peng, H.; Qin, L.; Qi, J.; Zuo, X.; Liu, J.-Y.; Zhang, J.-T. Determinants of 14-3-3σ protein dimerization and function in drug and radiation resistance. J. Biol. Chem. 2013, 288, 31447–31457. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-Y.; Li, Z.; Li, H.; Zhang, J.-T. Critical residue that promotes protein dimerization: A story of partially exposed Phe25 in 14-3-3σ. J. Chem. Inf. Model. 2011, 51, 2612–2625. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.; Chou, P.-C.; Velazquez-Torres, G.; Samudio, I.; Parreno, K.; Huang, Y.; Tseng, C.; Vu, T.; Gully, C.; Su, C.-H. The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming. Nat. Commun. 2015, 6, 7530. [Google Scholar] [CrossRef] [Green Version]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Ferguson, A.T.; Evron, E.; Umbricht, C.B.; Pandita, T.K.; Chan, T.A.; Hermeking, H.; Marks, J.R.; Lambers, A.R.; Futreal, P.A.; Stampfer, M.R. High frequency of hypermethylation at the 14-3-3 σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. USA 2000, 97, 6049–6054. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-H.; Lozano, G. Regulation of the p53-MDM2 pathway by 14-3-3 σ and other proteins. Semin. Cancer Biol. 2006, 16, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhao, R.; Lee, M.-H. 14-3-3σ, a p53 regulator, suppresses tumor growth of nasopharyngeal carcinoma. Mol. Cancer Ther. 2006, 5, 253–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.-Y.; Wen, Y.-Y.; Chen, C.-H.; Lozano, G.; Lee, M.-H. 14-3-3σ positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 2003, 23, 7096–7107. [Google Scholar] [CrossRef] [Green Version]
- West-Foyle, H.; Kothari, P.; Osborne, J.; Robinson, D.N. 14-3-3 proteins tune non-muscle myosin II assembly. J. Biol. Chem. 2018, 293, 6751–6761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, T.A.; Hermeking, H.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 1999, 401, 616–620. [Google Scholar] [CrossRef]
- Laronga, C.; Yang, H.-Y.; Neal, C.; Lee, M.-H. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J. Biol. Chem. 2000, 275, 23106–23112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.-Y.; Jeon, W.-K.; Bae, E.-J.; Kim, S.-T.; Lee, H.-J.; Kim, S.-J.; Kim, B.-C. 14-3-3 sigma and 14-3-3 zeta plays an opposite role in cell growth inhibition mediated by transforming growth factor-beta 1. Mol. Cells 2010, 29, 305–309. [Google Scholar] [CrossRef]
- Hong, H.-Y.; Jeon, W.-K.; Kim, S.-J.; Kim, B.-C. 14-3-3 σ is a new target up-regulated by transforming growth factor-β1 through a Smad3-dependent mechanism. Biochem. Biophys. Res. Commun. 2013, 432, 193–197. [Google Scholar] [CrossRef]
- Ingles-Esteve, J.; Morales, M.; Dalmases, A.; Garcia-Carbonell, R.; Jene-Sanz, A.; López-Bigas, N.; Iglesias, M.; Ruiz-Herguido, C.; Rovira, A.; Rojo, F. Inhibition of specific NF-κB activity contributes to the tumor suppressor function of 14-3-3σ in breast cancer. PLoS ONE 2012, 7, e38347. [Google Scholar] [CrossRef] [Green Version]
- Wolter, M.; de Vink, P.; Neves, J.F.; Srdanovic, S.; Higuchi, Y.; Kato, N.; Wilson, A.J.; Landrieu, I.; Brunsveld, L.; Ottmann, C. Selectivity via cooperativity: Preferential stabilization of the p65/14-3-3 interaction with semi-synthetic natural products. J. Am. Chem. Soc. 2020, 142, 11772–11783. [Google Scholar] [CrossRef]
- Aghazadeh, Y.; Papadopoulos, V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today 2016, 21, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Higuchi, Y.; Koschinsky, K.; Bartel, M.; Schumacher, B.; Thiel, P.; Nitta, H.; Preisig-Müller, R.; Schlichthörl, G.; Renigunta, V.; et al. A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface. Chem. Biol. 2013, 20, 583–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zúñiga, R.; Valenzuela, C.; Concha, G.; Brown, N.; Zúñiga, L. TASK-3 downregulation triggers cellular senescence and growth inhibition in breast cancer cell lines. Int. J. Mol. Sci. 2018, 19, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillory, X.; Wolter, M.; Leysen, S.; Neves, J.F.; Kuusk, A.; Genet, S.; Somsen, B.; Morrow, J.; Rivers, E.; van Beek, L. Fragment-based differential targeting of PPI stabilizer interfaces. J. Med. Chem. 2020, 63, 6694–6707. [Google Scholar] [CrossRef]
- Zhou, X.; Lei, Q.-Y. Regulation of TAZ in cancer. Protein Cell 2016, 7, 548–561. [Google Scholar] [CrossRef] [Green Version]
- Lodygin, D.; Yazdi, A.; Sander, C.; Herzinger, T.; Hermeking, H. Analysis of 14-3-3 expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene 2003, 22, 5519–5524. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, J.-Y.; Zhang, J.-T. 14-3-3σ, the double-edged sword of human cancers. Am. J. Transl. Res. 2009, 1, 326. [Google Scholar]
- Chan, S.Y.-Y.; To, K.-F.; Leung, S.-F.; Yip, W.W.-L.; Mak, M.K.-F.; Chung, G.T.-Y.; Lo, K.-W. 14-3-3σ expression as a prognostic marker in undifferentiated nasopharyngeal carcinoma. Oncol. Rep. 2010, 24, 949–955. [Google Scholar]
- Qi, Y.-J.; Wang, M.; Liu, R.-M.; Wei, H.; Chao, W.-X.; Zhang, T.; Lou, Q.; Li, X.-M.; Ma, J.; Zhu, H. Downregulation of 14-3-3σ correlates with multistage carcinogenesis and poor prognosis of esophageal squamous cell carcinoma. PLoS ONE 2014, 9, e95386. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Shao, Z.; Liu, J.; Zhan, W.; Gao, Q.; Pan, Z.; Wu, L.; Xu, L.; Ding, Y.; Zhao, L. COPS5 and LASP1 synergistically interact to downregulate 14-3-3σ expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int. J. Cancer 2018, 142, 1853–1864. [Google Scholar] [CrossRef]
- Umbricht, C.B.; Evron, E.; Gabrielson, E.; Ferguson, A.; Marks, J.; Sukumar, S. Hypermethylation of 14-3-3 σ (stratifin) is an early event in breast cancer. Oncogene 2001, 20, 3348–3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vercoutter-Edouart, A.-S.; Lemoine, J.; Le Bourhis, X.; Louis, H.; Boilly, B.; Nurcombe, V.; Révillion, F.; Peyrat, J.-P.; Hondermarck, H. Proteomic analysis reveals that 14-3-3σ is down-regulated in human breast cancer cells. Cancer Res. 2001, 61, 76–80. [Google Scholar] [PubMed]
- Urano, T.; Saito, T.; Tsukui, T.; Fujita, M.; Hosoi, T.; Muramatsu, M.; Ouchi, Y.; Inoue, S. Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature 2002, 417, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.H.; Gully, C.; Su, C.-H.; Velazquez-Torres, G.; Chou, P.-C.; Tseng, C.; Zhao, R.; Phan, L.; Shaiken, T.; Chen, J. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3σ. Oncogene 2011, 30, 4791. [Google Scholar] [CrossRef] [Green Version]
- Uchida, D.; Begum, N.; Almofti, A.; Kawamata, H.; Yoshida, H.; Sato, M. Frequent downregulation of 14-3-3 σ protein and hypermethylation of 14-3-3 σ gene in salivary gland adenoid cystic carcinoma. Br. J. Cancer 2004, 91, 1131–1138. [Google Scholar] [CrossRef] [Green Version]
- Yi, B.; Tan, S.X.; Tang, C.E.; Huang, W.G.; Cheng, A.L.; Li, C.; Zhang, P.F.; Li, M.Y.; Li, J.L.; Yi, H. Inactivation of 14-3-3 σ by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J. Cell. Biochem. 2009, 106, 858–866. [Google Scholar] [CrossRef]
- Sun, L.; Ain, Q.U.; Gao, Y.-S.; Khan, G.J.; Yuan, S.-t.; Roy, D. Effect of Marsdenia tenacissima extract on G2/M cell cycle arrest by upregulating 14-3-3σ and downregulating c-myc in vitro and in vivo. Chin. Herb. Med. 2019, 11, 169–176. [Google Scholar] [CrossRef]
- Han, B.; Xie, H.; Chen, Q.; Zhang, J.-T. Sensitizing hormone-refractory prostate cancer cells to drug treatment by targeting 14-3-3σ. Mol. Cancer Ther. 2006, 5, 903–912. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, H.; Han, B.; Zhang, J.-T. Identification of 14-3-3σ as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res. 2006, 66, 3248–3255. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-T.; Liu, Y. Use of comparative proteomics to identify potential resistance mechanisms in cancer treatment. Cancer Treat. Rev. 2007, 33, 741–756. [Google Scholar] [CrossRef] [Green Version]
- Ghahary, A.; Marcoux, Y.; Karimi-Busheri, F.; Li, Y.; Tredget, E.E.; Kilani, R.T.; Lam, E.; Weinfeld, M. Differentiated keratinocyte-releasable stratifin (14-3-3 sigma) stimulates MMP-1 expression in dermal fibroblasts. J. Investig. Dermatol. 2005, 124, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, P.; Roglin, L.; Meissner, N.; Hennig, S.; Kohlbacher, O.; Ottmann, C. Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions. Chem. Commun. (Camb.) 2013, 49, 8468–8470. [Google Scholar] [CrossRef] [PubMed]
- Corradi, V.; Mancini, M.; Santucci, M.; Carlomagno, T.; Sanfelice, D.; Mori, M.; Vignaroli, G.; Falchi, F.; Manetti, F.; Radi, M.; et al. Computational techniques are valuable tools for the discovery of protein-protein interaction inhibitors: The 14-3-3σ case. Bioorg. Med. Chem. Lett. 2011, 21, 6867–6871. [Google Scholar] [CrossRef] [PubMed]
- Iralde-Lorente, L.; Tassone, G.; Clementi, L.; Franci, L.; Munier, C.C.; Cau, Y.; Mori, M.; Chiariello, M.; Angelucci, A.; Perry, M.W. Identification of phosphate-containing compounds as new inhibitors of 14-3-3/c-Abl protein-protein interaction. ACS Chem. Biol. 2020, 15, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Stevers, L.M.; Sijbesma, E.; Botta, M.; MacKintosh, C.; Obsil, T.; Landrieu, I.; Cau, Y.; Wilson, A.J.; Karawajczyk, A.; Eickhoff, J. Modulators of 14-3-3 protein–protein interactions. J. Med. Chem. 2018, 61, 3755–3778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, A.M.; Hirsch, A.K.H. Molecular insight into specific 14-3-3 modulators: Inhibitors and stabilisers of protein–protein interactions of 14-3-3. Eur. J. Med. Chem. 2017, 136, 573–584. [Google Scholar] [CrossRef]
- Fullone, M. Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J. Biol. Chem. 1998, 273, 7698–7702. [Google Scholar] [CrossRef] [Green Version]
- Würtele, M.; Jelich-Ottmann, C.; Wittinghofer, A.; Oecking, C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003, 22, 987–994. [Google Scholar] [CrossRef] [Green Version]
- Doveston, R.G.; Kuusk, A.; Andrei, S.A.; Leysen, S.; Cao, Q.; Castaldi, M.P.; Hendricks, A.; Brunsveld, L.; Chen, H.; Boyd, H.; et al. Small-molecule stabilization of the p53—14-3-3 protein-protein interaction. FEBS Lett. 2017, 591, 2449–2457. [Google Scholar] [CrossRef] [Green Version]
- Andrei, S.A.; de Vink, P.; Sijbesma, E.; Han, L.; Brunsveld, L.; Kato, N.; Ottmann, C.; Higuchi, Y. Rationally designed semisynthetic natural product analogues for stabilization of 14-3-3 protein–protein interactions. Angew. Chem. Int. Ed. 2018, 57, 13470–13474. [Google Scholar] [CrossRef]
- Ottmann, C.; Wolter, M.; Valenti, D.; Cossar, P.J.; Levy, L.M.; Hristeva, S.; Genski, T.; Hoffmann, T.; Brunsveld, L.; Tzalis, D. Fragment based protein-protein interaction stabilizers via imine-based tethering. Angew. Chem. 2020, 59, 21520–21524. [Google Scholar]
- Ghaffari, A.; Li, Y.; Kilani, R.T.; Ghahary, A. 14-3-3σ associates with cell surface aminopeptidase N in the regulation of matrix metalloproteinase-1. J. Cell Sci. 2010, 123, 2996–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Cao, Z.; Xu, S.; Wang, W.; Wang, J. Revealing the binding modes and the unbinding of 14-3-3σ proteins and inhibitors by computational methods. Sci. Rep. 2015, 5, 16481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fokkens, M.; Schrader, T.; Klärner, F.-G. A Molecular tweezer for lysine and arginine. J. Am. Chem. Soc. 2005, 127, 14415–14421. [Google Scholar] [CrossRef] [PubMed]
- Mbarek, A.; Moussa, G.; Chain, J.L. Pharmaceutical applications of molecular tweezers, clefts and clips. Molecules 2019, 24, 1803. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Xu, D. Molecular dynamics investigations suggest a non-specific recognition strategy of 14-3-3sigma protein by tweezer: Implication for the inhibition mechanism. Front. Chem. 2019, 7, 237. [Google Scholar] [CrossRef] [Green Version]
- Bier, D.; Rose, R.; Bravo-Rodriguez, K.; Bartel, M.; Ramirez-Anguita, J.M.; Dutt, S.; Wilch, C.; Klärner, F.-G.; Sanchez-Garcia, E.; Schrader, T. Molecular tweezers modulate 14-3-3 protein–protein interactions. Nat. Chem. 2013, 5, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Corradi, V.; Mancini, M.; Manetti, F.; Petta, S.; Santucci, M.A.; Botta, M. Identification of the first non-peptidic small molecule inhibitor of the c-Abl/14-3-3 protein-protein interactions able to drive sensitive and Imatinib-resistant leukemia cells to apoptosis. Bioorg. Med. Chem. Lett. 2010, 20, 6133–6137. [Google Scholar] [CrossRef]
- Mancini, M.; Veljkovic, N.; Corradi, V.; Zuffa, E.; Corrado, P.; Pagnotta, E.; Martinelli, G.; Barbieri, E.; Santucci, M.A. 14-3-3 ligand prevents nuclear import of c-ABL protein in chronic myeloid leukemia. Traffic 2009, 10, 637–647. [Google Scholar] [CrossRef] [Green Version]
- Mancini, M.; Corradi, V.; Petta, S.; Barbieri, E.; Manetti, F.; Botta, M.; Santucci, M.A. A new nonpeptidic inhibitor of 14-3-3 induces apoptotic cell death in chronic myeloid leukemia sensitive or resistant to imatinib. J. Pharmacol. Exp. Ther. 2011, 336, 596–604. [Google Scholar] [CrossRef] [Green Version]
- Valensin, D.; Cau, Y.; Calandro, P.; Vignaroli, G.; Dello Iacono, L.; Chiariello, M.; Mori, M.; Botta, M. Molecular insights to the bioactive form of BV02, a reference inhibitor of 14-3-3sigma protein-protein interactions. Bioorg. Med. Chem. Lett. 2016, 26, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Iralde-Lorente, L.; Cau, Y.; Clementi, L.; Franci, L.; Tassone, G.; Valensin, D.; Mori, M.; Angelucci, A.; Chiariello, M.; Botta, M. Chemically stable inhibitors of 14-3-3 protein–protein interactions derived from BV02. J. Enzym. Inhib. Med. Chem. 2019, 34, 657–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Chung, B.; Ko, D.; Lim, H.-S. A solid-phase method for synthesis of dimeric and trimeric ligands: Identification of potent bivalent ligands of 14-3-3σ. Bioorg. Chem. 2019, 91, 103141. [Google Scholar] [CrossRef] [PubMed]
- Shi, M. Screening of 14-3-3σ natural product inhibitors by molecular docking. J. Adv. Phys. Chem. 2019, 8, 11–21. [Google Scholar] [CrossRef]





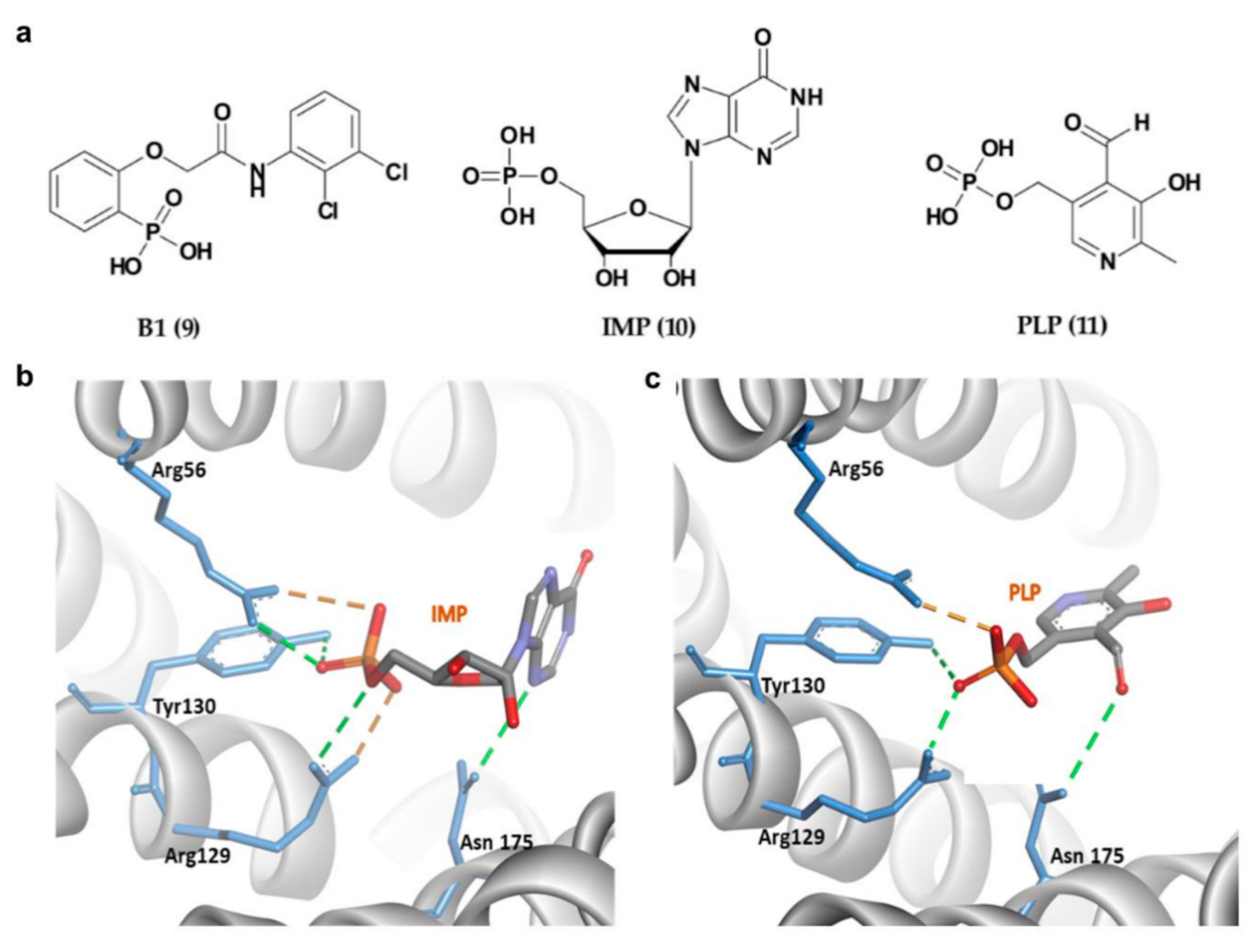


| No. | Chemical Classification | Mechanism | IC50/% Inhibition/LD50/KD | Main Residues Involved in the Interaction | References |
|---|---|---|---|---|---|
| 9 | Phosphonate-type inhibitors | 14-3-3σ/APN interaction disruption | IC50 of 81 ± 15 μM | Arg56, Arg129 and Tyr130, Arg60 | [94,104] |
| 10 | Phosphate-type inhibitors | 14-3-3σ/c-Abl interaction disruption | 34% at 1.5 mM | Arg56, Arg129 and Tyr130, Asn175 | [96] |
| 11 | 14-3-3σ/c-Abl interaction disruption | 74% at 1.5 mM | Arg56, Arg129 and Tyr130, Asn175 | [96] | |
| 12 | 14-3-3σ/C-Raf interaction disruption | 480 μM | Lys214 | [109] | |
| 14-3-3σ/ExoS interaction disruption | 520 μM | Lys214 | [109] | ||
| 13 | Carboxylate-type inhibitors | 14-3-3σ/c-Abl interaction disruption | LD50 = 1.04 μM | Lys49, Arg56, Arg60, Arg129 | [95,110,111,112,113,114] |
| 14 | 14-3-3σ/c-Abl interaction disruption | 5.2 ± 0.7 μM | Lys49, Arg56, Arg129, Tyr130, Asn175, Lys122 | ||
| 15 | 14-3-3σ/c-Abl interaction disruption | LD50 = 1.41 μM | Lys49, Arg56, Arg60, Arg129 | ||
| 16 | 14-3-3σ/c-Abl interaction disruption | 7.7 ± 2.0 μM | Arg56, Arg129, Lys49, Asn175, Lys122 | ||
| 17–20 | Peptide inhibitors | Disrupting 14-3-3σ interaction with its partners involved in cancer progression | KD 12.3 μM, KD = 59, 47, 55 nM | Targeting the two identical amphipathic grooves | [115] |
| 21–22 | Natural products | 14-3-3σ/partners interaction disruption | - | Targeting the amphipathic groove | [116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljabal, G.; Yap, B.K. 14-3-3σ and Its Modulators in Cancer. Pharmaceuticals 2020, 13, 441. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13120441
Aljabal G, Yap BK. 14-3-3σ and Its Modulators in Cancer. Pharmaceuticals. 2020; 13(12):441. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13120441
Chicago/Turabian StyleAljabal, Ghazi, and Beow Keat Yap. 2020. "14-3-3σ and Its Modulators in Cancer" Pharmaceuticals 13, no. 12: 441. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13120441





