The Effect of Novel Oleanolic Acid Oximes Conjugated with Indomethacin on the Nrf2-ARE And NF-κB Signaling Pathways in Normal Hepatocytes and Human Hepatocellular Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Spectral Characteristics of the Oleanolic Acid Oximes and Their Conjugates with Indomethacin
2.3. Effect of Indomethacin and Its Conjugates with OAO Derivatives on Cell Viability
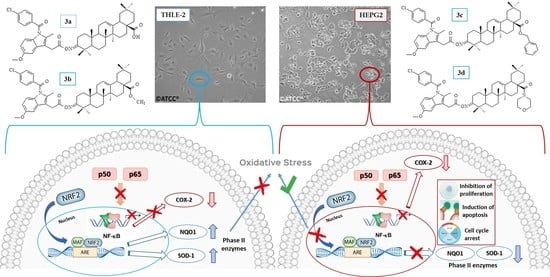
2.4. The Effect of Indomethacin and Its Conjugates with OAO Derivatives on Nrf2 Activation and Expression of Its Target Genes
2.5. The Effect of Indomethacin and Its Conjugates with OAO Derivatives on NF-κB Activation and Expression of COX-2 Gene
2.6. The Effect of Indomethacin and Its Conjugates with OAO Derivatives on the Nrf2 and NF-κB Subunits’ Transcript Levels in HepG2 Cells
2.7. The Effect of Indomethacin and Its Conjugates with OAO on Cell Cycle Distribution, Apoptosis, and Proliferation of HepG2 Cells
3. Materials and Methods
3.1. Chemistry
 ), 357.79 (6%, indomethacin), 406.34 (39%,
), 357.79 (6%, indomethacin), 406.34 (39%,  ), 790,41 (9%,
), 790,41 (9%,  ), 792.40 (5%,
), 792.40 (5%,  ), 808.42 (4%, M•+, molecular ion). Elem. anal.: for C49H61ClN2O6 calcd.: C: 72.71%, H: 7.60%, N: 3.46%, found: C: 72.73%, H: 7.59, %, N: 3.48%.
), 808.42 (4%, M•+, molecular ion). Elem. anal.: for C49H61ClN2O6 calcd.: C: 72.71%, H: 7.60%, N: 3.46%, found: C: 72.73%, H: 7.59, %, N: 3.48%. ), 357.79 (9%, indomethacin), 406.34 (34%,
), 357.79 (9%, indomethacin), 406.34 (34%,  ), 790,41 (13%,
), 790,41 (13%,  ), 746.43 (4%,
), 746.43 (4%,  ), 822.51 (5%, M•+, molecular ion). Elem. anal.: for C50H63ClN2O6 calcd.: C: 72.93%, H: 7.71%, N: 3.40%, found: C: 72.90%, H: 7.70, %, N: 3.42%.
), 822.51 (5%, M•+, molecular ion). Elem. anal.: for C50H63ClN2O6 calcd.: C: 72.93%, H: 7.71%, N: 3.40%, found: C: 72.90%, H: 7.70, %, N: 3.42%. ), 357.79 (7%, indomethacin), 406.34 (39%,
), 357.79 (7%, indomethacin), 406.34 (39%,  ), 790,41 (10%,
), 790,41 (10%,  ), 898.61 (2%, M•+, molecular ion). Elem. anal.: for C56H67ClN2O6 calcd.: C: 74.77%, H: 7.51%, N: 3.11%, found: C: 74.73%, H: 7.50, %, N: 3.12%.
), 898.61 (2%, M•+, molecular ion). Elem. anal.: for C56H67ClN2O6 calcd.: C: 74.77%, H: 7.51%, N: 3.11%, found: C: 74.73%, H: 7.50, %, N: 3.12%. ), 357.79 (6%, indomethacin), 406.34 (36%,
), 357.79 (6%, indomethacin), 406.34 (36%,  ), 790,41 (10%,
), 790,41 (10%,  ), 878.59.61 (3%, M•+, molecular ion). Elem. anal.: for C53H68ClN3O6 calcd.: C: 72.45%, H: 7.80%, N: 4.78%, found: C: 72.43%, H: 7.82, %, N: 3.76%.
), 878.59.61 (3%, M•+, molecular ion). Elem. anal.: for C53H68ClN3O6 calcd.: C: 72.45%, H: 7.80%, N: 4.78%, found: C: 72.43%, H: 7.82, %, N: 3.76%.3.2. Biological Assays
3.2.1. Cell Culture and Viability Assay
3.2.2. Nuclear and Cytosolic Fractions Preparation
3.2.3. Total RNA Isolation and cDNA Synthesis
3.2.4. Quantitative Real-Time PCR
3.2.5. Western Blot Analysis
3.2.6. Nrf2 and NF-κB Binding Assay
3.2.7. Cell Cycle Distribution
3.2.8. Apoptosis
3.2.9. Proliferation
3.2.10. Statistical Analysis
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Acknowledgments
Conflicts of Interest
References
- Ayeleso, T.B.; Matumba, M.G.; Mukwevho, E. Oleanolic acid and its derivatives: Biological activities and therapeutic potential in chronic diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farazi, P.A.; DePinho, R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer 2006, 9, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Husain, S.S.; Szabo, I.L.; Tarnawski, A.S. NSAID inhibition of GI cancer growth: Clinical implications and molecular mechanisms of action. Am. J. Gastroenterol. 2002, 97, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Li, Y.; Liu, X.; Deng, Q.; Yu, Y.; Yang, Z. Non-steroidal anti-inflammatory drugs, especially aspirin, are linked to lower risk and better survival of hepatocellular carcinoma: A meta-analysis. Cancer Manag. Res. 2018, 10, 2695–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meunier, L.; Larrey, D. Recent advances in hepatotoxicity of non-steroidal anti-inflammatory drugs. Ann. Hepatol. 2018, 17, 187–191. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Chattopadhyay, R.; Mitra, S.; Crowe, S.E. Oxidative stress: An essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol. Rev. 2014, 94, 329–354. [Google Scholar] [CrossRef] [Green Version]
- Juan-Reyes, N.S.; Gómez-Oliván, L.M.; Galar-Martínez, M.; Vieyra-Reyes, P.; García-Medina, S.; Islas-Flores, H.; Neri-Cruz, N. Effluent from an NSAID-manufacturing plant in Mexico induces oxidative stress on Cyprinus carpio. Water Air Soil Pollut. 2013, 224, 1689. [Google Scholar] [CrossRef]
- Sriuttha, P.; Sirichanchuen, B.; Permsuwan, U. Hepatotoxicity of non-steroidal anti-inflammatory drugs: A systematic review of randomized controlled trials. Int. J. Hepatol. 2018, 2018, 5253623. [Google Scholar] [CrossRef]
- Kakisaka, K.; Yoshida, Y.; Suzuki, Y.; Sato, T.; Kuroda, H.; Miyasaka, A.; Takikawa, Y. Serum markers for mitochondrial dysfunction and cell death are possible predictive indicators for drug-induced liver injury by direct acting antivirals. Hepatol. Res. 2018, 48, 78–86. [Google Scholar] [CrossRef] [Green Version]
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 Talks, Who’s Listening? Antioxid. Redox Signal. 2010, 13, 1649–1663. [Google Scholar] [CrossRef] [Green Version]
- Krajka-Kuźniak, V.; Bednarczyk-Cwynar, B.; Paluszczak, J.; Szaefer, H.; Narożna, M.; Zaprutko, L.; Baer-Dubowska, W. Oleanolic acid oxime derivatives and their conjugates with aspirin modulate the NF-κB-mediated transcription in HepG2 hepatoma cells. Bioorg. Chem. 2019, 93, 103326. [Google Scholar] [CrossRef] [PubMed]
- Crook, D.; Collins, A.J. Comparison of effects of aspirin and indomethacin on human platelet prostaglandin synthetase. Ann. Rheum. Dis. 1977, 36, 459–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blobaum, A.L.; Uddin, J.; Felts, A.S.; Crews, B.C.; Rouzer, C.A.; Marnett, L.J. The 2′-Trifluoromethyl analogue of indomethacin is a potent and selective COX-2 inhibitor. ACS Med. Chem. Lett. 2013, 4, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk-Cwynar, B.; Zaprutko, L.; Froelich, A. Beckmann rearrangement of oxime obtained from oleanolic acid. Structure elucidation of the initial oxime. J. Mol. Struct. 2013, 1053, 115–121. [Google Scholar] [CrossRef]
- Bednarczyk-Cwynar, B.; Wachowiak, N.; Szulc, M.; Kamińska, E.; Bogacz, A.; Bartkowiak-Wieczorek, J.; Zaprutko, L.; Mikolajczak, P.L. Strong and long-lasting antinociceptive and anti-inflammatory conjugate of naturally occurring oleanolic acid and aspirin. Front. Pharmacol. 2016, 7, 202. [Google Scholar] [CrossRef] [Green Version]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Bobkiewicz-Kozlowska, T.; Zaprutko, L. Oleanolic acid a-lactams inhibit the growth of HeLa, KB, MCF-7 and Hep-G2 cancer cell lines at micromolar concentrations. Anti Cancer Agents Med. Chem. 2016, 16, 579–592. [Google Scholar] [CrossRef]
- Kaminskyy, D.; Bednarczyk-Cwynar, B.; Vasylenko, O.; Kazakova, O.; Zimenkovsky, B.; Zaprutko, L.; Lesyk, R. Synthesis of new potential anticancer agents based on 4-thiazolidinone and oleanane scaffolds. Med. Chem. Res. 2012, 21, 3568–3580. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. The Nrf2-ARE signaling pathway: An update on its regulation and possible role in cancer prevention and treatment. Pharmacol. Rep. 2017, 69, 393–402. [Google Scholar] [CrossRef]
- Tsui, R.; Kearns, J.; Lynch, C.; Vu, D.; Ngo, K.A.; Basak, S.; Ghosh, G.; Hoffmann, A. IκBβ enhances the generation of the low-affinity NF-κB/RelA homodimer. Nat. Commun. 2015, 6, 7068. [Google Scholar] [CrossRef] [Green Version]
- Sun, S. Non-canonical NF-κB signaling pathway. Cell Res. 2011, 21, 71–85. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, M.L.; Baeuerle, P.A. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991, 10, 3805–3817. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Wan, Y.; Huang, C. The biological functions of NF-κB1 (p50) and its potential as an anti-cancer target. Curr. Cancer Drug Targets 2009, 9, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, D.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Motohashi, H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015, 88, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Raghunath, A.; Sundarraj, K.; Arfuso, F.; Sethi, G.; Perumal, E. Dysregulation of Nrf2 in hepatocellular carcinoma: Role in cancer progression and chemoresistance. Cancers 2018, 10, 481. [Google Scholar] [CrossRef] [Green Version]









| Compound | THLE-2 | HepG2 |
|---|---|---|
| IND | >150 | >150 |
| 3a | 79.5 ± 2.25 | 33.5 ± 4.61 |
| 3b | 44 ± 1.22 | 34 ± 2.5 |
| 3c | 41.5 ± 3.5 | 40 ± 5.03 |
| 3d | 42.5 ± 3.6 | 38.5 ± 5.48 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| NRF2 | 5′ATTGCTACTAATCAGGCTCAG | 5′GTTTGGCTTCTGGACTTGG |
| SOD-1 | 5′CGACAGAAGGAAAGTAATG | 5′TGGATAGAGGATTAAAGTGAGG |
| NQO1 | 5′CAATTCAGAGTGGCATTC | 5′GAAGTTTAGGTCAAAGAGG |
| NF-κB p50 | 5′ATCATCCACCTTCATTCTCAA | 5′AATCCTCCACCACATCTTCC |
| NF-κB p65 | 5′CGCCTGTCCTTTCTCATC | 5′ACCTCAATGTCCTCTTTCTG |
| COX-2 | 5′CCTGTGCCTGATGATTGC | 5′CAGCCCGTTGGTGAAAGC |
| PBGD | 5′TCAGATAGCATACAAGAGACC | 5′TGGAATGTTACGAGCAGTG |
| TBP | 5′GGCACCACTCCACTGTATC | 5′GGGATTATATTCGGCGTTTCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narożna, M.; Krajka-Kuźniak, V.; Bednarczyk-Cwynar, B.; Kleszcz, R.; Baer-Dubowska, W. The Effect of Novel Oleanolic Acid Oximes Conjugated with Indomethacin on the Nrf2-ARE And NF-κB Signaling Pathways in Normal Hepatocytes and Human Hepatocellular Cancer Cells. Pharmaceuticals 2021, 14, 32. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14010032
Narożna M, Krajka-Kuźniak V, Bednarczyk-Cwynar B, Kleszcz R, Baer-Dubowska W. The Effect of Novel Oleanolic Acid Oximes Conjugated with Indomethacin on the Nrf2-ARE And NF-κB Signaling Pathways in Normal Hepatocytes and Human Hepatocellular Cancer Cells. Pharmaceuticals. 2021; 14(1):32. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14010032
Chicago/Turabian StyleNarożna, Maria, Violetta Krajka-Kuźniak, Barbara Bednarczyk-Cwynar, Robert Kleszcz, and Wanda Baer-Dubowska. 2021. "The Effect of Novel Oleanolic Acid Oximes Conjugated with Indomethacin on the Nrf2-ARE And NF-κB Signaling Pathways in Normal Hepatocytes and Human Hepatocellular Cancer Cells" Pharmaceuticals 14, no. 1: 32. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14010032