Dicopper(II)-EDTA Chelate as a Bicephalic Receptor Model for a Synthetic Adenine Nucleoside
Abstract
:1. Introduction
2. Results and Discussion
2.1. Comments Concerning the Synthesis
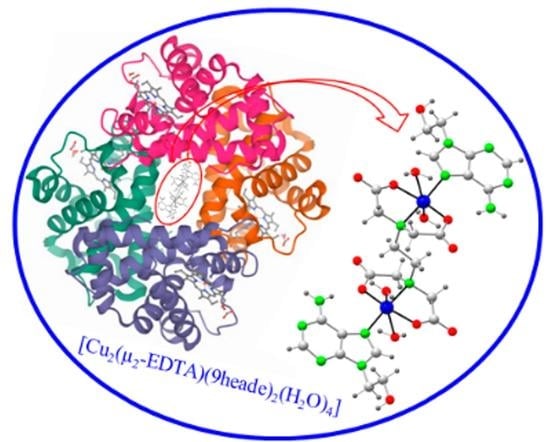
2.2. Molecular and Crystal Structure
2.3. Computational Results
2.4. Thermal Stability
2.5. Electron Spin Resonance (ESR) Spectra and Magnetic Properties
2.6. Remarks on the Suitability of the Cu2(µ-EDTA) Chelate as Bicephalic Receptor for 9heade
3. Materials and Methods
3.1. Synthesis of the Compound [Cu2(µ2-EDTA)(9heade)2(H2O)4]·3H2O (1)
3.2. Crystal Structure Determination
3.3. Computational Methods
3.4. Other Physical Measurements
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Choquesillo-Lazarte, D.; Brandi-Blanco, M.P.; García-Santos, I.; González-Pérez, J.M.; Castiñeiras, A.; Niclós-Gutiérrez, J. Interligand Interactions Involved in the Molecular Recognition between Copper (II) Complexes and Adenine or Related Purines. Coord. Chem. Rev. 2008, 252, 1241–1256. [Google Scholar] [CrossRef]
- Verma, S.; Mishra, A.K.; Kumar, J. The Many Facets of Adenine: Coordination, Crystal Patterns, and Catalysis. Acc. Chem. Res. 2010, 43, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.K.; Domínguez-Martín, A.; Brandi-Blanco, M.P.; Choquesillo-Lazarte, D.; Nurchi, V.M.; Niclós-Gutiérrez, J. Metal Ion Binding Modes of Hypoxanthine and Xanthine Versus the Versatile Behaviour of Adenine. Coord. Chem. Rev. 2012, 256, 193–221. [Google Scholar] [CrossRef]
- Domínguez-Martín, A.; Brandi-Blanco, M.P.; Matilla-Hernández, A.; El Bakkali, H.; Nurchi, V.M.; González-Pérez, J.M.; Castiñeiras, A.; Niclós-Gutiérrez, J. Unravelling the Versatile Metal Binding Modes of Adenine: Looking at the Molecular Recognition Patterns of Deaza-and Aza-Adenines in Mixed Ligand Metal Complexes. Coord. Chem. Rev. 2013, 257, 2814–2838. [Google Scholar] [CrossRef]
- Domínguez-Martín, A.; Choquesillo-Lazarte, D.; González-Pérez, J.M.; Castiñeiras, A.; Niclós-Gutiérrez, J. Molecular Recognition Patterns of 2-Aminopurine Versus Adenine: A View Through Ternary Copper (II) Complexes. J. Inorg. Biochem. 2011, 105, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Rojas-González, P.X.; Castiñeiras, A.; González-Pérez, J.M.; Choquesillo-Lazarte, D.; Niclós-Gutiérrez, J. Interligand Interactions Controlling the µ-N7,N9-Metal Bonding of Adenine (AdeH) to the N-Benzyliminodiacetato(2-) Copper(II) Chelate and Promoting the N9 Versus N3 Tautomeric Proton Transfer: Molecular and Crystal Structure of [Cu2(NBzIDA)2(H2O)2(µ-N7,N9-Ade(N3)H)]·3H2O. Inorg. Chem. 2002, 41, 6190–6192. [Google Scholar] [CrossRef] [PubMed]
- Stevanović, N.L.; Aleksic, I.; Kljun, J.; Skaro Bogojevic, S.; Veselinovic, A.; Nikodinovic-Runic, J.; Turel, I.; Djuran, M.I.; Glišić, B.Đ. Copper (II) and Zinc (II) Complexes with the Clinically Used Fluconazole: Comparison of Antifungal Activity and Therapeutic Potential. Pharmaceuticals 2021, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Doniz Kettenmann, S.; Nossol, Y.; Louka, F.R.; Legrande, J.R.; Marine, E.; Fischer, R.C.; Mautner, F.A.; Hergl, V.; Kulak, N.; Massoud, S.S. Copper (II) Complexes with Tetradentate Piperazine-Based Ligands: DNA Cleavage and Cytotoxicity. Inorganics 2021, 9, 12. [Google Scholar] [CrossRef]
- Padnya, P.; Shibaeva, K.; Arsenyev, M.; Baryshnikova, S.; Terenteva, O.; Shiabiev, I.; Khannanov, A.; Boldyrev, A.; Gerasimov, A.; Grishaev, D.; et al. Catechol-Containing Schiff Bases on Thiacalixarene: Synthesis, Copper (II) Recognition, and Formation of Organic-Inorganic Copper-Based Materials. Molecules 2021, 26, 2334. [Google Scholar] [CrossRef] [PubMed]
- Shelman, S.E.; Gibson, D.; Wang, A.H.-J.; Lippard, S. Crystal and Molecular Structure of Cis-[Pt(NH3)2[d(pGpG)]], the Principal Adduct Formed by Cis-Diamminedichloroplatinum (II) with DNA. J. Am. Chem. Soc. 1988, 110, 7368–7381. [Google Scholar] [CrossRef]
- Maldonado, N.; Amo-Ochoa, P. The Role of Coordination Compounds in Virus Research. Different Approaches and Rrends. Dalton Trans. 2021, 50, 2310–2323. [Google Scholar] [CrossRef] [PubMed]
- Velo-Gala, I.; Barceló-Oliver, M.; Gil, D.M.; González-Pérez, J.M.; Castiñeiras, A.; Domínguez-Martín, A. Deciphering the H-Bonding Preference on Nucleoside Molecular Recognition Through Model Copper (II) Compounds. Pharmaceuticals 2021, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, N.; García-Rubiño, M.E.; Domínguez-Martín, A.; Choquesillo-Lazarte, D.; Franconetti, A.; Frontera, A.; Castiñeiras, A.; González-Pérez, J.M.; Niclós-Gutiérrez, J. Molecular and Supra-Molecular Recognition Patterns in Ternary Copper (II) or Zinc (II) Complexes with Selected Rigid-Planar Chelators and a Synthetic Adenine-Nucleoside. J. Inorg. Biochem. 2020, 203, 110920. [Google Scholar] [CrossRef]
- Belmont-Sánchez, J.C.; García-Rubiño, M.E.; Frontera, A.; Matilla-Hernández, A.; Castiñeiras, A.; Niclós-Gutiérrez, J. Novel Cd (II) Coordination Polymers Afforded with EDTA or Trans-1,2-CDTA Chelators and Imidazole, Adenine, or 9-(2-hydroxyethyl) Adenine Coligands. Crystals 2020, 10, 391. [Google Scholar] [CrossRef]
- Sushrutha, S.R.; Hota, R.; Natarajan, S. Adenine-Based Coordination Polymers: Synthesis, Structure, and Properties. Eur. J. Inorg. Chem. 2016, 2016, 2962–2964. [Google Scholar] [CrossRef]
- Hammud, H.H.; Travis Holman, K.; Al-Noaimi, M.; Sadiq Sheikh, N.; Ghannoum, A.M.; Bouhadir, K.H.; Masoud, M.S.; Karnati, R.K. Structures of Selected Transition Metal Complexes with 9-(2-Hydroxyethyl) Adenine: Potentiometric Complexation and DFT Studies. J. Mol. Struct. 2020, 1205, 127548. [Google Scholar] [CrossRef]
- Fecher, R.; Boswell, K.H.; Wittick, J.J.; Shen, T.Y. Nucleosides VI: The Synthesis and Optical Properties of the 5’-adenin-9yl)-5’-deoxy Derivatives of the Thymidine and 2’-deoxyadenosine. Carbohydr. Res. 1970, 13, 105–111. [Google Scholar] [CrossRef]
- Xia, R.; Sun, L.; Qu, G. Microwave-Assisted Synthesis of 9-(2-Hydroxyetyl) Adenine as Intermediate of Adefovir Dipivoxyl. Jingxi Shiyou Huagong 2016, 33, 74–78. [Google Scholar]
- Kim, J.C.; Jung, J.; Rho, Y.; Kim, M.; Kwon, W.; Kim, H.; Kim, I.J.; Kim, J.R.; Ree, M. Well-Defined DNA-Mimic Brush Polymers Bearing Adenine Moieties: Synthesis, Layer-by-Layer Self-Assembly, and Biocompatibility. Biomacromolecules 2011, 12, 2822–2833. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, A.; Shibata, M.; Sasada, Y. Three Crystalline Forms of 9-(2-Hydroxyethyl) Adenine Resulting from the Different Stacking of Hydrogen-Bonded Layers. Acta Crystallogr. 1986, C42, 13036–13040. [Google Scholar] [CrossRef]
- Kitade, Y.; Hayashi, M.; Yatome, C.; Ghajima, M.; Nagase, H. Inhibitory Effect on the HT-1080 Tumor Cell Invasion in Vitro Using 9-(2’-hydroxyethyl) Adenine 2’-Phosphates. Bioorg. Med. Chem. Lett. 1997, 7, 833–836. [Google Scholar] [CrossRef]
- Polynova, T.N.; Dvorstnova, N.V.; Fillippova, T.V.; Porai-Khosits, M.A.; Nekrasov, Y.V. X-ray Structure Study of Ammonium μ-Hexamethylenediaminetetraacetato-Bis-(Trioxomolybdate) Dihydrate. Koord. Khim. 1986, 12, 1484–1487. [Google Scholar]
- Antsyshkina, A.S.; Sadikov, G.G.; Poznyak, A.L.; Segienko, V.S. Crystal Structures of [Cu2(Edta)(Py)2(H2O)2]·2H2O and [Cu(Him)6]{Cu(Im)4[Cu(Edta)(Im)]2}·6H2O, Products of the Interaction of (Ethylenediaminetetraacetato) Diaquadicopper (II) with Pyridine and Imidazole. Russ. J. Inorg. Chem. 2006, 51, 241–252. [Google Scholar] [CrossRef]
- Deng, Z.-P.; Gao, S.; Huo, L.-H.; Zhao, H. Synthesis, crystal structure and thermal stability of “Windmill-shaped” dinuclear cadmium complex [Cd-2 (3-PyOH)(4)(FBA)(4)]. J. Inorg. Chem. 2007, 23, 1089–1092. [Google Scholar]
- Belmont-Sánchez, J.C.; García-Rubiño, M.E.; Frontera, A.; González-Pérez, J.M.; Castiñeiras, A.; Niclós-Gutiérrez, J. H-Bonds, π-Stacking and (Water) OH/π Interactions in (μ4-EDTA) Bis (Imidazole) Dicopper (II) Dehydrate. Crystals 2021, 11, 48. [Google Scholar] [CrossRef]
- Huheey, J.E.; Keiter, E.A.; Keiter, R.L. Norganic Chemistry. In Principles of Structure and Reactivity; HarperCollins College Press: New York, NY, USA, 1993; Chapter 9; ISBN1 10:006042995X. ISBN2 13:9780060429959. [Google Scholar]
- Kaim, W.; Schwedeski, B.; Klein, A. Bioinorganic Chemistry-Inorganic Elements in the Chemistry of Life, 2nd ed.; Wiley: Hoboken, NJ, USA, 2013; ISBN 978-0-470-97523-7. [Google Scholar]
- Hathaway, B.J. Comprehensive Coordination Chemistry; Pergamon Press: Brighton, Oxford, UK, 1987; Volume 5, pp. 533–773. [Google Scholar]
- Hathaway, B.J.; Billing, D.E. The Electronic Properties and Stereochemistry of Mono-Nuclear Complexes of the Copper (II) Ion. Coord. Chem. Rev. 1970, 5, 143–278. [Google Scholar] [CrossRef]
- Bleaney, B.; Bowers, K.D. Anomalous Paramagnetism of Copper Acetate. Proc. R. Soc. London Ser. A 1952, 214, 451–465. [Google Scholar] [CrossRef]
- Bruker. Apex3 v2019.1-0, SAINT V8.40A; Bruker AXS Inc.: Madison, WI, USA, 2019. [Google Scholar]
- Sheldrick, G.M. Program for Empirical Absorption Correction of Area Detector Data. SADABS 1997. [Google Scholar]
- Sheldrick, G.M. A Short Hstory of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Wilson, A.J.C. International Tables for Crystallography; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; Volume C. [Google Scholar]
- Spek, A.L. Structure Validation in Chemical Crystallography. Acta Crystallogr. 2009, D65, 148–155. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Revision A.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A Consistent and Accurate ab Initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154122. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.M.N.; Dutta, D.; Verma, A.K.; Nath, H.; Frontera, A.; Sharma, P.; Bhattacharyya, M.K. Antiproliferative Evaluation and Supramolecular Association Involving Electrostatically Enhanced π-π Interaction in Isostructural Coordination Solids of Mn (II), Co (II) and Zn (II) Chlorobenzoates: Experimental and Theoretical Studies. Inorg. Chim. Acta 2019, 498, 119161–119174. [Google Scholar] [CrossRef]
- Bader, R.F.W. A Bond Path: A Universal Indicator of Bonded Interactions. J. Phys. Chem. 1998, 102, 7314–7323. [Google Scholar] [CrossRef]
- Keith, T.A. AIMAll (Version 19.02.13); TK Gristmill Software: Overland Park, KS, USA, 2019; Available online: aim.tkgristmill.com (accessed on 28 April 2021).













| Atoms | Distance/Angle |
|---|---|
| Cu(1)-O(21) | 1.943(4) |
| Cu(1)-O (11) | 1.978(4) |
| Cu(1)-N(7) | 2.032(6) |
| Cu(1)-N(10) | 2.039(5) |
| Cu(1)-O(1) | 2.331(5) |
| Cu(1)-O(2) | 2.816(6) |
| O(21)-Cu(1)-O(11) | 164.4(2) |
| N(7)-Cu(1)-N(10) | 174.8(2) |
| O(1)-Cu(1)-O(2) | 163.9(2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Rubiño, M.E.; Matilla-Hernández, A.; Frontera, A.; Lezama, L.; Niclós-Gutiérrez, J.; Choquesillo-Lazarte, D. Dicopper(II)-EDTA Chelate as a Bicephalic Receptor Model for a Synthetic Adenine Nucleoside. Pharmaceuticals 2021, 14, 426. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14050426
García-Rubiño ME, Matilla-Hernández A, Frontera A, Lezama L, Niclós-Gutiérrez J, Choquesillo-Lazarte D. Dicopper(II)-EDTA Chelate as a Bicephalic Receptor Model for a Synthetic Adenine Nucleoside. Pharmaceuticals. 2021; 14(5):426. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14050426
Chicago/Turabian StyleGarcía-Rubiño, María Eugenia, Antonio Matilla-Hernández, Antonio Frontera, Luis Lezama, Juan Niclós-Gutiérrez, and Duane Choquesillo-Lazarte. 2021. "Dicopper(II)-EDTA Chelate as a Bicephalic Receptor Model for a Synthetic Adenine Nucleoside" Pharmaceuticals 14, no. 5: 426. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14050426