Therapeutic Potential of Glycosyl Flavonoids as Anti-Coronaviral Agents
Abstract
:1. Introduction
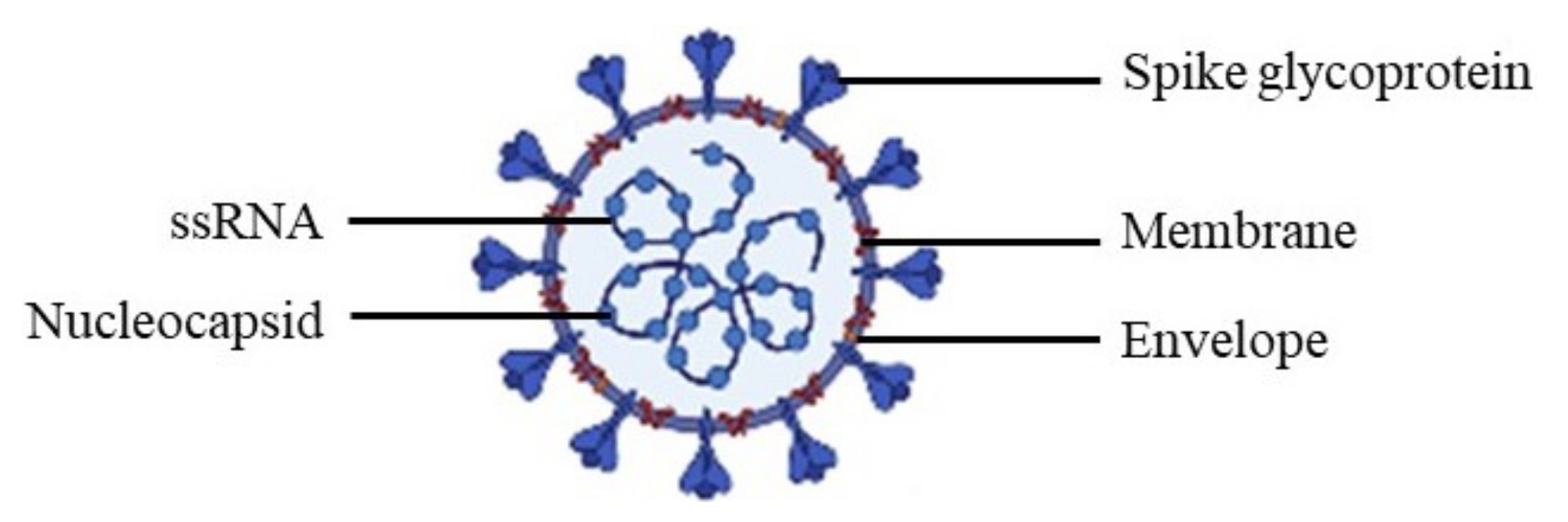
2. Severe Acute Respiratory Syndrome Coronavirus 2
2.1. Spike Glycoprotein
2.2. 3C-Like Protease and Papain-Like Protease
2.3. RNA-Dependent RNA Polymerase
3. Structure and Function of Glycosyl Flavonoids
3.1. Biological Activity of Glycosyl Flavonoids
3.2. Antiviral Activity of Glycosyl Flavonoids
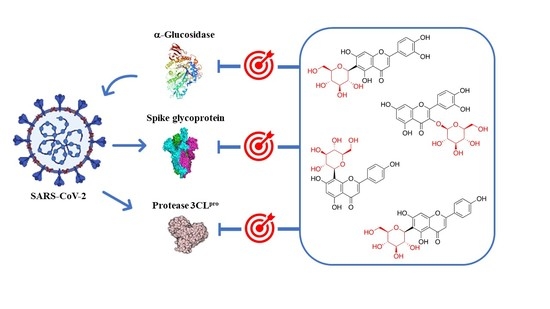
4. Glycosyl Flavonoids as Anti-Coronaviral Agents
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, P.; Lu, X.; Xu, C.; Sun, W.; Pan, B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020, 92, 548–551. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Capanoglu, E.; Jassbi, A.R.; Miron, A. Advance on the Flavonoid C-glycosides and Health Benefits. Crit. Rev. Food Sci. Nutr. 2016, 56, S29–S45. [Google Scholar] [CrossRef]
- Forkmann, G.; Martens, S. Metabolic engineering and applications of flavonoids. Curr. Opin. Biotechnol. 2001, 12, 155–160. [Google Scholar] [CrossRef]
- Ferreyra, M.L.E.; Rius, S.P.; Ecasati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [Green Version]
- Courts, F.L.; Williamson, G. The Occurrence, Fate and Biological Activities of C-glycosyl Flavonoids in the Human Diet. Crit. Rev. Food Sci. Nutr. 2015, 55, 1352–1367. [Google Scholar] [CrossRef] [Green Version]
- Sak, K. Cytotoxicity of dietary flavonoids on different human cancer types. Pharmacogn. Rev. 2014, 8, 122–146. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J. Dietary Flavonoid Aglycones and Their Glycosides: Which Show Better Biological Significance? Crit. Rev. Food Sci. Nutr. 2015, 57, 1874–1905. [Google Scholar] [CrossRef] [PubMed]
- Pietta, P.-G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Rice-Evans, C. Flavonoid Antioxidants. Curr. Med. Chem. 2001, 8, 797–807. [Google Scholar] [CrossRef]
- Ribeiro, D.; Freitas, M.; Tomé, S.M.; Silva, A.M.; Porto, G.; Fernandes, E. Modulation of human neutrophils’ oxidative burst by flavonoids. Eur. J. Med. Chem. 2013, 67, 280–292. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Braidy, N.; Habtemariam, S.; Orhan, I.E.; Daglia, M.; Manayi, A.; Gortzi, O. Neuroprotective effects of chrysin: From chemistry to medicine. Neurochem. Int. 2015, 90, 224–231. [Google Scholar] [CrossRef]
- Nakajima, A.; Ohizumi, Y. Potential Benefits of Nobiletin, A Citrus Flavonoid, against Alzheimer’s Disease and Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batra, P.; Sharma, A.K. Anti-cancer potential of flavonoids: Recent trends and future perspectives. 3 Biotech 2013, 3, 439–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abotaleb, M.; Samuel, S.M.; Varghese, E.; Varghese, S.; Kubatka, P.; Líšková, A.; Büsselberg, D. Flavonoids in Cancer and Apoptosis. Cancers 2018, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Proença, C.; Freitas, M.; Ribeiro, D.; Tomé, S.M.; Oliveira, E.F.T.; Viegas, M.F.; Araújo, A.N.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A.; et al. Evaluation of a flavonoids library for inhibition of pancreatic α-amylase towards a structure–activity relationship. J. Enzym. Inhib. Med. Chem. 2019, 34, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Kaleem, M.; Ahmed, Z.; Shafiq, H. Therapeutic potential of flavonoids and their mechanism of action against microbial and viral infections—A review. Food Res. Int. 2015, 77, 221–235. [Google Scholar] [CrossRef]
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: Promising natural compounds against viral infections. Arch. Virol. 2017, 162, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Liu, H.; Li, J.; Yao, X.; Chen, Y.; Ke, C.; Liu, S. Structure-activity relationship of flavonoid bifunctional inhibitors against Zika virus infection. Biochem. Pharmacol. 2020, 177, 113962. [Google Scholar] [CrossRef]
- Rengasamy, K.R.; Khan, H.; Gowrishankar, S.; Lagoa, R.J.; Mahomoodally, F.M.; Khan, Z.; Suroowan, S.; Tewari, D.; Zengin, G.; Hassan, S.T.; et al. The role of flavonoids in autoimmune diseases: Therapeutic updates. Pharmacol. Ther. 2019, 194, 107–131. [Google Scholar] [CrossRef]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus that Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Genet. 2009, 7, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Kochi, A.N.; Tagliari, A.P.; Forleo, G.B.; Fassini, G.M.; Tondo, C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020, 31, 1003–1008. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.R. COVID-19 and the nervous system. J. Neurovirol. 2020, 26, 143–148. [Google Scholar] [CrossRef]
- Hess, D.C.; Eldahshan, W.; Rutkowski, E. COVID-19-Related Stroke. Transl. Stroke Res. 2020, 11, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Williams, S.J.; Goddard-Borger, E.D. α-Glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem. Soc. Trans. 2020, 48, 1287–1295. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nat. Cell Biol. 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601. [Google Scholar] [CrossRef] [PubMed]
- Konwar, M.; Sarma, D. Advances in developing small molecule SARS 3CLpro inhibitors as potential remedy for corona virus infection. Tetrahedron 2021, 77, 131761. [Google Scholar] [CrossRef] [PubMed]
- Mouffouk, C.; Mouffouk, S.; Mouffouk, S.; Hambaba, L.; Haba, H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). Eur. J. Pharmacol. 2021, 891, 173759. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Progress in Developing Inhibitors of SARS-CoV-2 3C-Like Protease. Microorganisms 2020, 8, 1250. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, S.; Teng, T.; Abdalla, A.; Zhu, W.; Xie, L.; Wang, Y.; Guo, X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020, 12, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdusalam, A.A.A.; Murugaiyah, V. Identification of Potential Inhibitors of 3CL Protease of SARS-CoV-2 From ZINC Database by Molecular Docking-Based Virtual Screening. Front. Mol. Biosci. 2020, 7, 603037. [Google Scholar] [CrossRef]
- Kuo, C.-J.; Chao, T.-L.; Kao, H.-C.; Tsai, Y.-M.; Liu, Y.-K.; Wang, L.H.-C.; Hsieh, M.-C.; Chang, S.-Y.; Liang, P.-H. Kinetic Characterization and Inhibitor Screening for the Proteases Leading to Identification of Drugs against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65, 02577-20. [Google Scholar] [CrossRef] [PubMed]
- Freitas, B.T.; Durie, I.A.; Murray, J.; Longo, J.E.; Miller, H.C.; Crich, D.; Hogan, R.J.; Tripp, R.A.; Pegan, S.D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020, 6, 2099–2109. [Google Scholar] [CrossRef]
- Adhikari, N.; Baidya, S.K.; Saha, A.; Jha, T. Structural Insight into the Viral 3C-Like Protease Inhibitors: Comparative SAR/QSAR Approaches. In Viral Proteases and Their Inhibitors; Elsevier BV: Amsterdam, The Netherlands, 2017; pp. 317–409. [Google Scholar]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Yang, H.; Xie, W.; Xue, X.; Yang, K.; Ma, J.; Liang, W.; Zhao, Q.; Zhou, Z.; Pei, D.; Ziebuhr, J.; et al. Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases. PLoS Biol. 2005, 3, e324. [Google Scholar] [CrossRef]
- Báez-Santos, Y.M.; John, S.E.S.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Spatial and temporal roles of SARS-CoV PLpro —A snapshot. FASEB J. 2021, 35, e21197. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 575, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Lenschow, D.J.; Lai, C.; Frias-Staheli, N.; Giannakopoulos, N.V.; Lutz, A.; Wolff, T.; Osiak, A.; Levine, B.; Schmidt, R.E.; García-Sastre, A.; et al. From the cover: IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 2007, 104, 1371–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picarazzi, F.; Vicenti, I.; Saladini, F.; Zazzi, M.; Mori, M. Targeting the RdRp of Emerging RNA Viruses: The Structure-Based Drug Design Challenge. Molecules 2020, 25, 5695. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ahn, D.-G.; Choi, J.-K.; Taylor, D.R.; Oh, J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [Green Version]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.; Decroly, E.; Snijder, E.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Li, B.; Qiao, M.; Li, J.; Xu, H.; Zhang, L.; Zhang, X. Advances on the in vivo and in vitro glycosylations of flavonoids. Appl. Microbiol. Biotechnol. 2020, 104, 6587–6600. [Google Scholar] [CrossRef] [PubMed]
- Plaza, M.; Pozzo, T.; Liu, J.; Ara, K.Z.G.; Turner, C.; Karlsson, E.N. Substituent Effects on in Vitro Antioxidizing Properties, Stability, and Solubility in Flavonoids. J. Agric. Food Chem. 2014, 62, 3321–3333. [Google Scholar] [CrossRef] [PubMed]
- Slámová, K.; Kapešová, J.; Valentová, K. “Sweet Flavonoids”: Glycosidase-Catalyzed Modifications. Int. J. Mol. Sci. 2018, 19, 2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
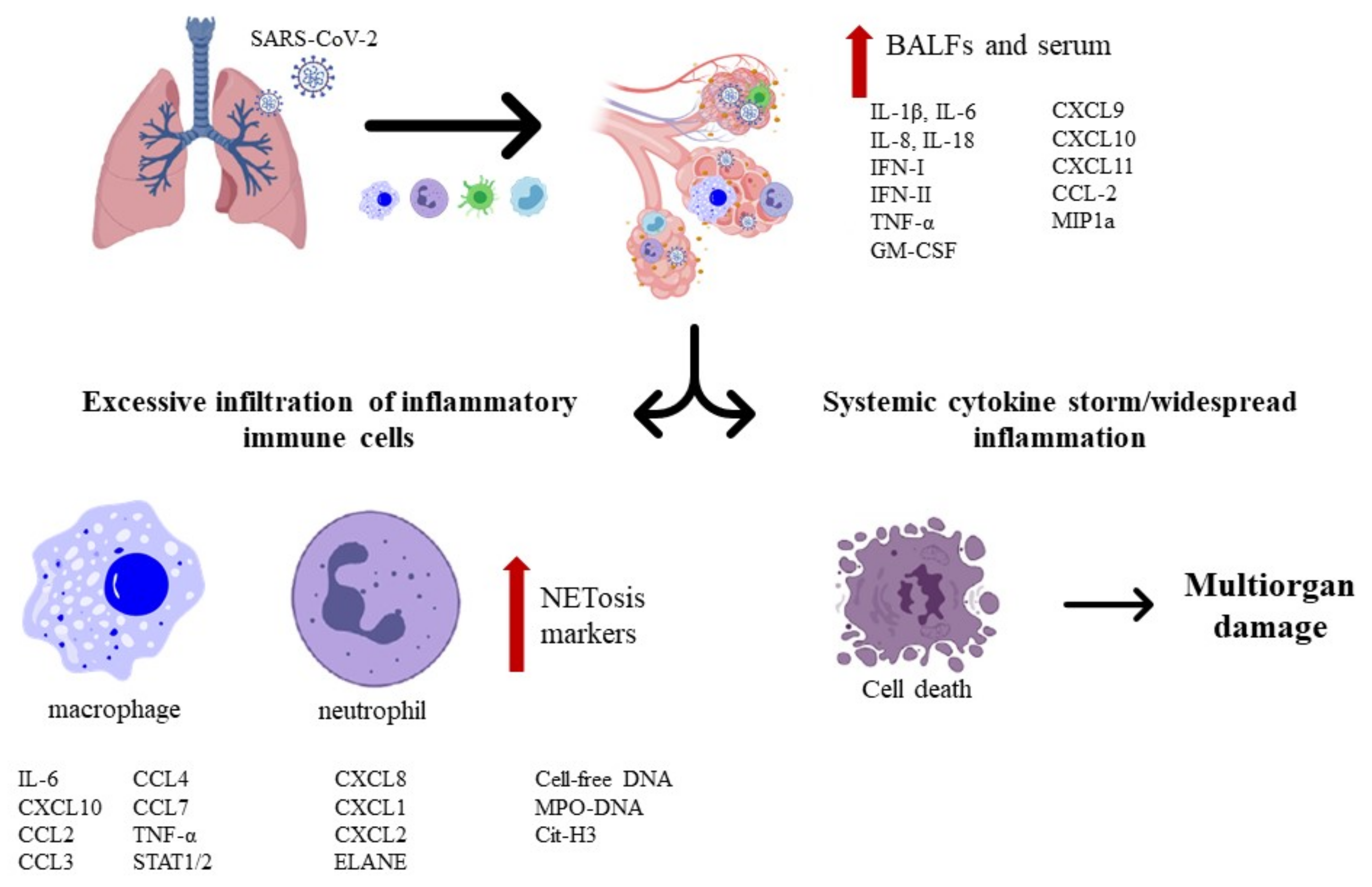
- Beltrán-García, J.; Osca-Verdegal, R.; Pallardó, F.V.; Ferreres, J.; Rodríguez, M.; Mulet, S.; Carbonell, F.; García-Giménez, J.L. Oxidative Stress and Inflammation in COVID-19-Associated Sepsis: The Potential Role of Anti-Oxidant Therapy in Avoiding Disease Progression. Antioxidants 2020, 9, 936. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lou, J.; Luo, C.; Zhou, L.; Wang, M.; Wang, L. Phenolic Compounds from Halimodendron halodendron (Pall.) Voss and Their Antimicrobial and Antioxidant Activities. Int. J. Mol. Sci. 2012, 13, 11349–11364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.-J.; Tai, B.H.; Cuong, N.M.; Kim, Y.-H.; Jang, H.-D. Antioxidative and anti-inflammatory effect of quercetin and its glycosides isolated from mampat (Cratoxylum formosum). Food Sci. Biotechnol. 2012, 21, 587–595. [Google Scholar] [CrossRef]
- Parhiz, H.; Roohbakhsh, A.; Soltani, F.; Rezaee, R.; Iranshahi, M. Antioxidant and Anti-Inflammatory Properties of the Citrus Flavonoids Hesperidin and Hesperetin: An Updated Review of their Molecular Mechanisms and Experimental Models. Phytother. Res. 2015, 29, 323–331. [Google Scholar] [CrossRef]
- Liskova, A.; Samec, M.; Koklesova, L.; Samuel, S.M.; Zhai, K.; Al-Ishaq, R.K.; Abotaleb, M.; Nosal, V.; Kajo, K.; Ashrafizadeh, M.; et al. Flavonoids against the SARS-CoV-2 induced inflammatory storm. Biomed. Pharmacother. 2021, 138, 111430. [Google Scholar] [CrossRef]
- Wunpathe, C.; Potue, P.; Maneesai, P.; Bunbupha, S.; Prachaney, P.; Kukongviriyapan, U.; Kukongviriyapan, V.; Pakdeechote, P. Hesperidin Suppresses Renin-Angiotensin System Mediated NOX2 Over-Expression and Sympathoexcitation in 2K-1C Hypertensive Rats. Am. J. Chin. Med. 2018, 46, 751–767. [Google Scholar] [CrossRef]
- Agarwal, O.P. The anti-inflammatory action of nepitrin, a flavonoid. Inflamm. Res. 1982, 12, 298–302. [Google Scholar] [CrossRef]
- Jin, X.; Liu, M.; Zhang, D.; Zhong, X.; Du, K.; Qian, P.; Yao, W.; Gao, H.; Wei, M. Baicalin mitigates cognitive impairment and protects neurons from microglia-mediated neuroinflammation via suppressing NLRP 3 inflammasomes and TLR 4/ NF -κB signaling pathway. CNS Neurosci. Ther. 2019, 25, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jing, Y.-Y.; Zeng, C.-Y.; Li, C.-G.; Xu, L.-H.; Yan, L.; Bai, W.-J.; Zha, Q.-B.; Ouyang, D.-Y.; He, X.-H. Scutellarin Suppresses NLRP3 Inflammasome Activation in Macrophages and Protects Mice against Bacterial Sepsis. Front. Pharmacol. 2018, 8, 975. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Block, T.M.; Guo, J.-T. Antiviral therapies targeting host ER alpha-glucosidases: Current status and future directions. Antivir. Res. 2013, 99, 251–260. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kwon, C.-S.; Son, K.H. Inhibition of Alpha-glucosidase and Amylase by Luteolin, a Flavonoid. Biosci. Biotechnol. Biochem. 2000, 64, 2458–2461. [Google Scholar] [CrossRef] [PubMed]
- Shibano, M.; Kakutani, K.; Taniguchi, M.; Yasuda, M.; Baba, K. Antioxidant constituents in the dayflower (Commelina communis L.) and their α-glucosidase-inhibitory activity. J. Nat. Med. 2008, 62, 349–353. [Google Scholar] [CrossRef]
- Choo, C.; Sulong, N.; Man, F.; Wong, T. Vitexin and isovitexin from the Leaves of Ficus deltoidea with in-vivo α-glucosidase inhibition. J. Ethnopharmacol. 2012, 142, 776–781. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Li, Q.; Bi, K.-S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-K.; Xia, Y.; Yang, Z.-Y.; Natschke, S.L.M.; Lee, K.-H. Recent Advances in the Discovery and Development of Flavonoids and their Analogues as Antitumor and Anti-HIV Agents. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1998; Volume 439, pp. 191–225. [Google Scholar]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef]
- Lalani, S.; Poh, C.L. Flavonoids as Antiviral Agents for Enterovirus A71 (EV-A71). Viruses 2020, 12, 184. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Kroeker, A.; He, S.; Kozak, R.; Audet, J.; Mbikay, M.; Chrétien, M. Prophylactic Efficacy of Quercetin 3-β-O-D-Glucoside against Ebola Virus Infection. Antimicrob. Agents Chemother. 2016, 60, 5182–5188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.J.; Song, J.H.; Park, K.S.; Kwon, D.H. Inhibitory effects of quercetin 3-rhamnoside on influenza A virus replication. Eur. J. Pharm. Sci. 2009, 37, 329–333. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Chang, Y.-C.; Hsiao, N.-W.; Hsieh, J.-L.; Wang, C.-Y.; Kung, S.-H.; Tsai, F.-J.; Lan, Y.-C.; Lin, C.-W. Fisetin and rutin as 3C protease inhibitors of enterovirus A71. J. Virol. Methods 2012, 182, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, P.; Chen, X.; Wang, W.; Jin, Y. Saururus chinensis (Lour.) Baill blocks enterovirus 71 infection by hijacking MEK1–ERK signaling pathway. Antivir. Res. 2015, 119, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Q.; Fu, T.; Dongyan, Y.; Mikovits, J.A.; Ruscetti, F.W.; Wang, J.M. Flavonoid Baicalin Inhibits HIV-1 Infection at the Level of Viral Entry. Biochem. Biophys. Res. Commun. 2000, 276, 534–538. [Google Scholar] [CrossRef]
- Chu, M.; Xu, L.; Zhang, M.-B.; Chu, Z.-Y.; Wang, Y.-D. Role of Baicalin in Anti-Influenza Virus A as a Potent Inducer of IFN-Gamma. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, Y.; Wu, T.; Jin, Y.; Cheng, J.; Wan, C.; Qian, W.; Xing, F.; Shi, W. The Antiviral Effect of Baicalin on Enterovirus 71 In Vitro. Viruses 2015, 7, 4756–4771. [Google Scholar] [CrossRef] [Green Version]
- Tsai, F.-J.; Lin, C.-W.; Lai, C.-C.; Lan, Y.-C.; Lai, C.-H.; Hung, C.-H.; Hsueh, K.-C.; Lin, T.-H.; Chang, H.C.; Wan, L.; et al. Kaempferol inhibits enterovirus 71 replication and internal ribosome entry site (IRES) activity through FUBP and HNRP proteins. Food Chem. 2011, 128, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Gunaseelan, S.; Wong, K.Z.; Min, N.; Sun, J.; Ismail, N.K.B.M.; Tan, Y.J.; Lee, R.C.H.; Chu, J.J.H. Prunin suppresses viral IRES activity and is a potential candidate for treating enterovirus A71 infection. Sci. Transl. Med. 2019, 11, eaar5759. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.-W.; Geng, C.-A.; Jiang, F.-Q.; Ma, Y.-B.; He, K.; Zhou, N.-J.; Zhang, X.-M.; Zhou, J.; Chen, J.-J. Chemical constituents of Swertia yunnanensis and their anti-hepatitis B virus activity. Fitoterapia 2013, 89, 175–182. [Google Scholar] [CrossRef]
- Yarmolinsky, L.; Huleihel, M.; Zaccai, M.; Ben-Shabat, S. Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia 2012, 83, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.T.; Suárez, A.I.; Serrano, M.L.; Baptista, J.; Pujol, F.H.; Rangel, H.R. The role of the glycosyl moiety of myricetin derivatives in anti-HIV-1 activity in vitro. AIDS Res. Ther. 2017, 14, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.-Q.; Wang, R.; Yang, X.; Shang, M.; Ma, C.; Shoyama, Y. Antiviral Flavonoid-Type C-Glycosides from the Flowers of Trollius chinensis. Chem. Biodivers. 2006, 3, 343–348. [Google Scholar] [CrossRef]
- Nayak, M.K.; Agrawal, A.S.; Bose, S.; Naskar, S.; Bhowmick, R.; Chakrabarti, S.; Sarkar, S.; Chawla-Sarkar, M. Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 2014, 69, 1298–1310. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tao, L.; Xu, H. Chinese herbal medicines as a source of molecules with anti-enterovirus 71 activity. Chin. Med. 2016, 11, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Tao, G.; Liu, J.; Cai, J.; Huang, Z.; Chen, J.-X. Current Prevention of COVID-19: Natural Products and Herbal Medicine. Front. Pharmacol. 2020, 11, 588508. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Leung, E.L.-H.; Liu, X.; Liu, K.; Wang, Q.; Lan, Y.; Li, X.; Yu, H.; Cui, L.; et al. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19). Pharmacol. Res. 2020, 158, 104929. [Google Scholar] [CrossRef]
- Yang, R.; Liu, H.; Bai, C.; Wang, Y.; Zhang, X.; Guo, R.; Wu, S.; Wang, J.; Leung, E.; Chang, H.; et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): In silico and experimental study. Pharmacol. Res. 2020, 157, 104820. [Google Scholar] [CrossRef]
- Solnier, J.; Fladerer, J.-P. Flavonoids: A complementary approach to conventional therapy of COVID-19? Phytochem. Rev. 2020, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ngwa, W.; Kumar, R.; Thompson, D.; Lyerly, W.; Moore, R.; Reid, T.-E.; Lowe, H.; Toyang, N. Potential of Flavonoid-Inspired Phytomedicines against COVID-19. Molecules 2020, 25, 2707. [Google Scholar] [CrossRef] [PubMed]
- Diniz, L.R.L.; Filho, C.D.S.M.B.; Fielding, B.C.; De Sousa, D.P. Natural Antioxidants: A Review of Studies on Human and Animal Coronavirus. Oxidative Med. Cell. Longev. 2020, 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.H.; Woo, H.-J.; Kang, H.-K.; Nguyen, V.D.; Kim, Y.-M.; Kim, D.-W.; Ahn, S.-A.; Xia, Y.; Kim, D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012, 34, 831–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, S.; Kim, H.; Kim, S.; Shin, D.H.; Kim, M. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019, 94, 2023–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-R.; Wang, Y.-C.; Lin, Y.W.; Chou, S.-Y.; Chen, S.-F.; Liu, L.T.; Wu, Y.-T.; Kuo, C.-J.; Chen, T.S.-S.; Juang, S.-H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorganic Med. Chem. Lett. 2005, 15, 3058–3062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huitema, C.; Niu, C.; Yin, J.; James, M.N.; Eltis, L.D.; Vederas, J.C. Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase. Bioorg. Chem. 2008, 36, 229–240. [Google Scholar] [CrossRef]
- Shitrit, A.; Zaidman, D.; Kalid, O.; Bloch, I.; Doron, D.; Yarnizky, T.; Buch, I.; Segev, I.; Ben-Zeev, E.; Segev, E.; et al. Conserved interactions required for inhibition of the main protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Su, H.; Zhou, F.; Huang, Z.; Ma, X.; Natarajan, K.; Zhang, M.; Huang, Y.; Su, H. Molecular Insights into Small-Molecule Drug Discovery for SARS-CoV-2. Angew. Chem. Int. Ed. 2021, 60, 9789–9802. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Kim, D.Y.; Kim, M.-S.; Shin, D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzym. Inhib. Med. Chem. 2020, 35, 1539–1544. [Google Scholar] [CrossRef]
- Dubey, K.; Dubey, R. Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomed. J. 2020, 43, 363–367. [Google Scholar] [CrossRef]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velazquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef]
- Rizzuti, B.; Grande, F.; Conforti, F.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Ortega-Alarcon, D.; Vega, S.; Reyburn, H.T.; Abian, O.; Velazquez-Campoy, A. Rutin is a low micromolar inhibitor of SARS-CoV-2 main protease 3CLpro: Implications for drug design of quercetin analogs. Biomedicines 2021, 9, 375. [Google Scholar] [CrossRef]
- Cherrak, S.A.; Merzouk, H.; Mokhtari-Soulimane, N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS ONE 2020, 15, e0240653. [Google Scholar] [CrossRef]
- A Da Silva, F.M.; Da Silva, K.P.A.; De Oliveira, L.P.M.; Costa, E.V.; Koolen, H.H.; Pinheiro, M.L.B.; De Souza, A.Q.L.; De Souza, A.D.L. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp). Memórias do Instituto Oswaldo Cruz 2020, 115, 200207. [Google Scholar] [CrossRef]
- Agrawal, P.K.; Agrawal, C.; Blunden, G. Rutin: A potential antiviral for repurposing as a SARS-CoV-2 Main Protease (Mpro) inhibitor. Nat. Prod. Commun. 2021, 16, 1–12. [Google Scholar] [CrossRef]
- Sharma, A.; Goyal, S.; Yadav, A.K.; Kumar, P.; Gupta, L. In-silico screening of plant-derived antivirals against main protease, 3CLpro and endoribonuclease, NSP15 proteins of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 1–15. [Google Scholar] [CrossRef]
- Pandey, P.; Rane, J.S.; Chatterjee, A.; Kumar, A.; Khan, R.; Prakash, A.; Ray, S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: An in silico study for drug development. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 732–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, P.; Khan, F.; Rana, A.K.; Srivastava, Y.; Jha, S.K.; Jha, N.K. A Drug Repurposing Approach Towards Elucidating the Potential of Flavonoids as COVID-19 Spike Protein Inhibitors. Biointerface Res. Appl. Chem. 2020, 11, 8482–8501. [Google Scholar] [CrossRef]
- Jain, A.S.; Sushma, P.; Dharmashekar, C.; Beelagi, M.S.; Prasad, S.K.; Shivamallu, C.; Prasad, A.; Syed, A.; Marraiki, N.; Prasad, K.S. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J. Biol. Sci. 2021, 28, 1040–1051. [Google Scholar] [CrossRef]
- Biagioli, M.; Marchianò, S.; Roselli, R.; Di Giorgio, C.; Bellini, R.; Bordoni, M.; Gidari, A.; Sabbatini, S.; Francisci, D.; Fiorillo, B.; et al. Discovery of a AHR pelargonidin agonist that counter-regulates ACE2 expression and attenuates ACE2-SARS-CoV-2 interaction. Biochem. Pharmacol. 2021, 188, 114564. [Google Scholar] [CrossRef] [PubMed]
- Guler, H.I.; Ay Sal, F.; Can, Z.; Kara, Y.; Yildiz, O.; Belduz, A.O.; Çanakci, S.; Kolayli, S. Targeting CoV-2 Spike RBD and ACE-2 interaction with flavonoids of anatolian propolis by in silico and in vitro studies in terms of possible COVID-19 therapeutics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rameshkumar, M.R.; Indu, P.; Arunagirinathan, N.; Venkatadri, B.; El-Serehy, H.A.; Ahmad, A. Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins: A molecular docking study. Saudi J. Biol. Sci. 2021, 28, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, S.; Kumar, H.D.V.; Nandan, M.; Mantesh, M.; Shankarappa, K.S.; Venkataravanappa, V.; Basha, C.R.J.; Reddy, C.N.L. In silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in Ayurveda medicine in inhibiting SARS-CoV-2. 3 Biotech 2021, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]



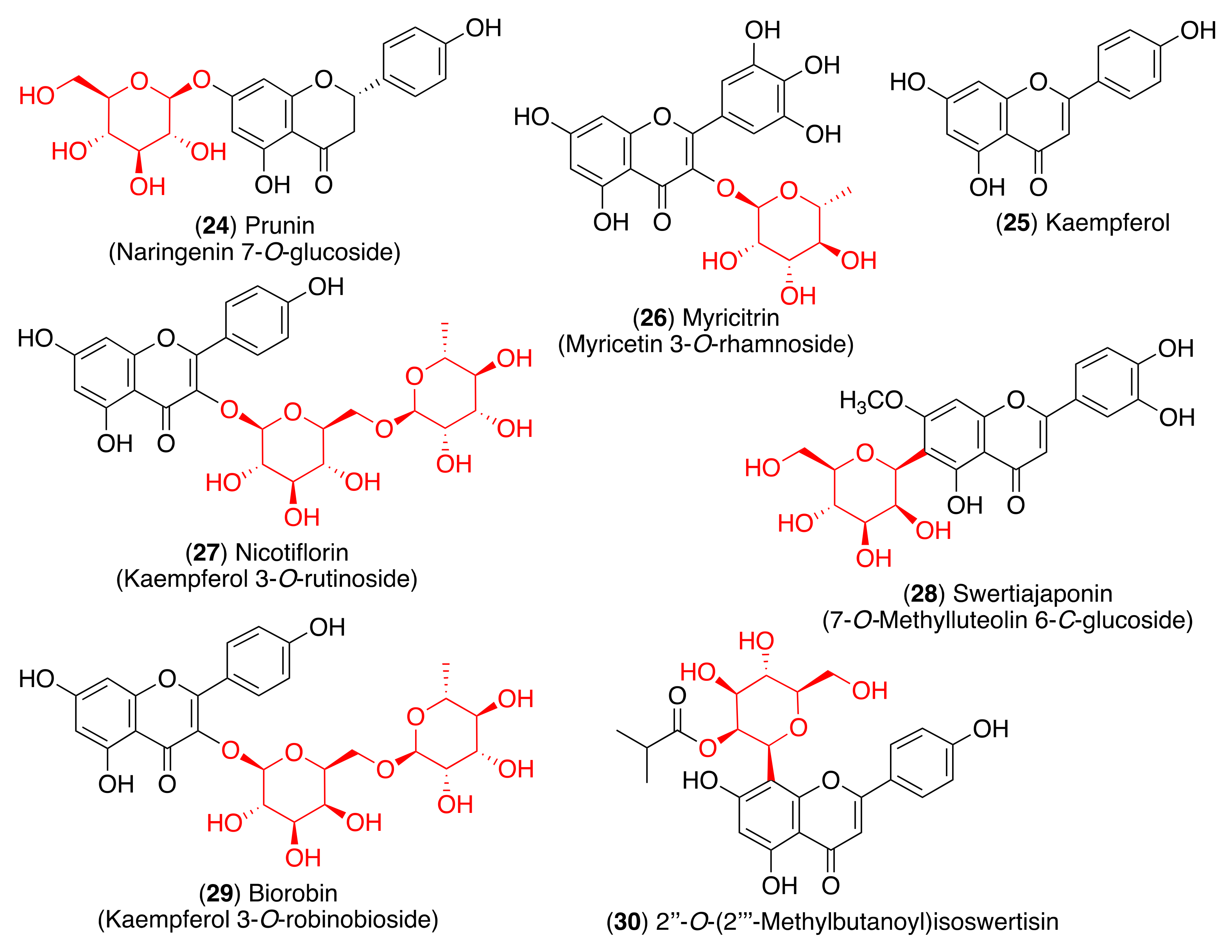
| Virus | Compound | IC50 (μM) | Reference |
|---|---|---|---|
| Ebola virus | Isoquercitrin (6) | 5.3 | [81] |
| Enterovirus A71 | Baicalin (10) | 11.1 | [87] |
| Hesperidin (8) | - a | [88] | |
| Prunin (24) | - a | [89] | |
| Rutin (13) | 109.6 | [83] | |
| Hepatitis B virus | Isovitexin (23) | - a | [90] |
| Herpes simplex virus | Rutin (13) | - a | [91] |
| Nicotiflorin (27) | - a | [91] | |
| Biorobin (29) | - a | [91] | |
| Human immunodeficiency virus type 1 | Baicalin (10) | - a | [85] |
| Myricitrin (26) | 7.9 | [92] | |
| Influenza A virus | 2″-O-(2‴-Methylbutanoyl) isoswertisin (30) | 147.8 | [93] |
| Quercitrin (7) | - a | [82] | |
| Baicalin (10) | - a | [86,94] | |
| Respiratory syncytial virus | Isoorientin (20) | 12.7 | [95] |
| Swertisin (22) | 112.0 | [95] | |
| Isovitexin (23) | 23.1 | [95] | |
| Swertiajaponin (28) | 27.1 | [95] |
| Target | Compound | IC50 (μM) | Reference |
|---|---|---|---|
| MERS-CoV 3CLpro | Isoquercitrin (6) | - a | [103] |
| SARS-CoV 3CLpro | Pectolinarin (14) | 37.78 | [104] |
| Rhoifolin (18) | 27.45 | [104] | |
| SARS-CoV-2 3CLpro | Baicalin (10) | 6.41 34.71 | [105] [110] |
| Pectolinarin (14) | 51.64 | [110] |
| Target | Compound | Amino Acid Residues | Reference |
|---|---|---|---|
| SARS-CoV-2 3CLpro | Narcissoside (4) | Arg188, Glu166, His164, Cys145, Asn14, Cys44, His41, Gln192, Thr190 | [111] |
| Rutin (13) | Leu141, Thr26, Cys145, His41, Met49, Tyr54, Met165, Glu166 | [113,114,115,116] | |
| Quercitrin (7) | Thr26, Phe140, Leu141, Gly143, His163, Arg188, Met49, Cys145 | [114] | |
| Nicotiflorin (27) | Met49, Met165, Glu166, Thr190, Cys145, His41 | [115] | |
| Myricetin 3-O-rutinoside (31) | Tyr54, His41, Met49, Met165, Thr26, Cys145, Ser144, Leu141, Gly143, Asn142, His163, Glu166 | [114] | |
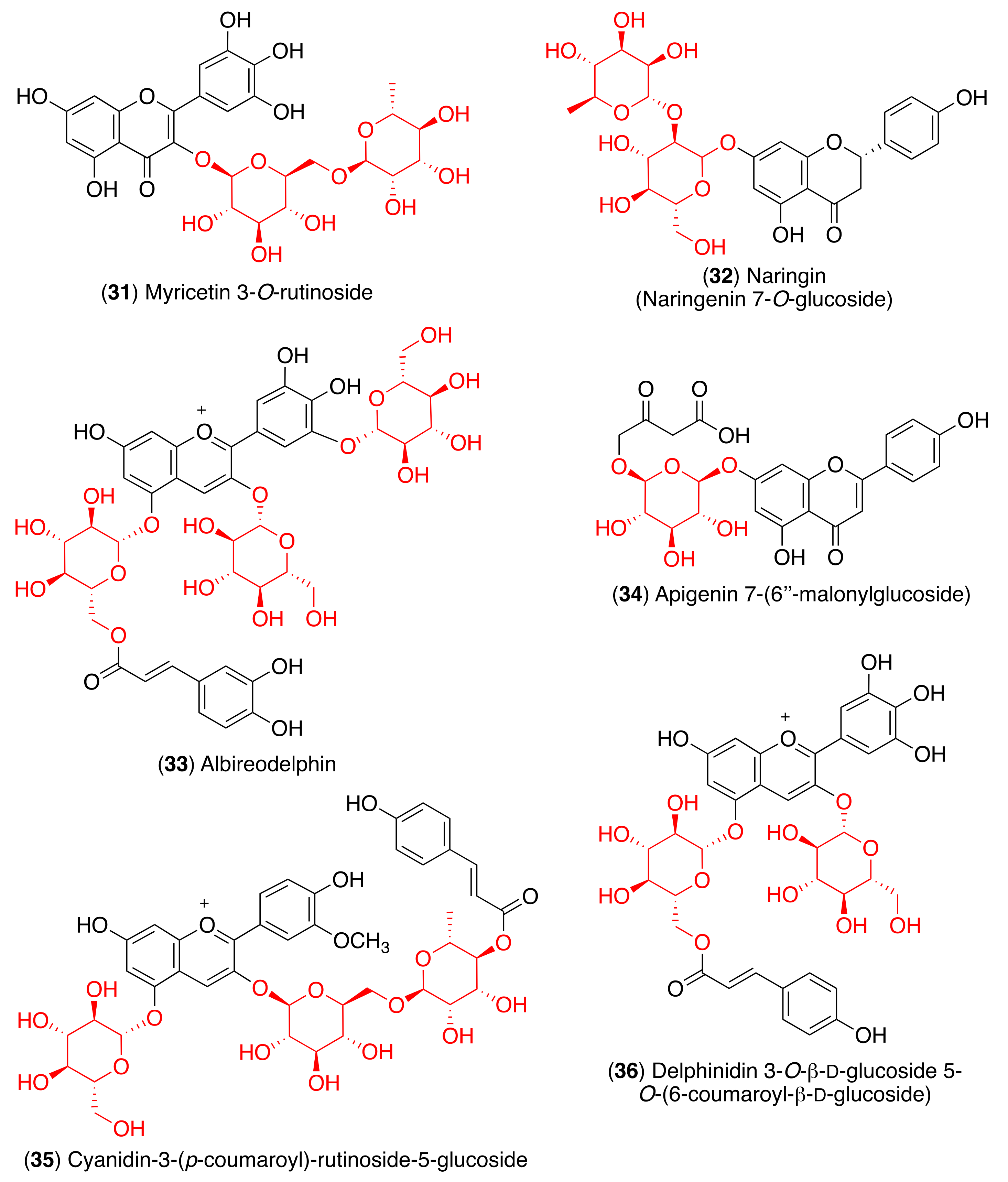
| Albireodelphin (33) | Lys5, Val125, Lys137, Ser139, Thr199, Glu288 | [124] | |
| Apigenin 7-(6″-malonylglucoside) (34) | Gln110, Thr111, Thr292, Asp295 | [124] | |
| Cyanidin-3-(p-coumaroyl)-rutinoside-5-glucoside (35) | Thr111, Gln110, Asn151 | [124] | |
| Delphinidin 3-O-β-d-glucoside 5-O-(6-coumaroyl-β-d-glucoside) (36) | Asp153, Asn151, Ser158, Thr111, Ile249 | [124] | |
| Isoquercitrin (6) | Leu141, His163, Met165 | [125] | |
| Spike glycoprotein | Baicalin (10) | Lys964, Gln965, Leu962, Thr961, Ser1003, Ala958, Tyr1007, Gln1011, Gln1010, Agr1014 | [120] |
| Naringin (32) | Asn290, Ile291, His374, Leu370, Leu410, Ala413, Pro415, Phe438, Gln442, Asp367, Thr371, Lys441, Glu406, Ser409 | [121] | |
| Albireodelphin (33) | Lys621, Asp623, Phe793, Lys798, Asp760, Asp761, Trp800, Glu811, Cys813, Ser814 | [124] | |
| Apigenin 7-(6″-malonylglucoside) (34) | Asp452, Trp617, Tyr619, Lys621, Asp760, Asp761, Trp800 | [124] | |
| Cyanidin-3-(p-coumaroyl)-rutinoside-5-glucoside (35) | Asp164, Ile548, Ser549, Arg553, Arg555, Asp760, Asp761 | [124] | |
| Delphinidin 3-O-β-d-glucoside 5-O-(6-coumaroyl-β-d-glucoside) (36) | Ser549, Arg553, Arg555, Thr556, Cys622, Asp623, Asp760, Asp761 | [124] | |
| Isoquercitrin (6) | Thr998, Arg995, Asp994 | [125] | |
| RdRp | Rutin (13) | Tyr455, Arg553, Ala554, Asp452, Arg624, Asp623, Asn691, Ser759, Thr556, Asp760, Cys622 | [115] |
| Albireodelphin (33) | Arg348, Asp350, His378, Asp382, Phe390, Asn394, Asn397, Glu398, His401, Glu402, Arg514 | [124] | |
| Apigenin 7-(6″-malonylglucoside) (34) | Als348, Asp350, His378, Asp382, Gly395, Asn394, His401 | [124] | |
| Cyanidin-3-(p-coumaroyl)-rutinoside-5-glucoside (35) | Ser44, Asp206, Als348, Asp350, Asn397, Glu398, Ser511, Arg514 | [124] | |
| Delphinidin 3-O-β-d-glucoside 5-O-(6-coumaroyl-β-d-glucoside) (36) | Try127, His345, Ala348, Asp350, Asp382, Tyr385, Asn394, Asn397, Arg401, His505, Arg514, Tyr515 | [124] | |
| Isoquercitrin (6) | Ala125, His133 | [125] | |
| SARS-CoV-2 PLpro | Isoquercitrin (6) | His74, Arg83, Tyr155, Asn157, His176 | [125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godinho, P.I.C.; Soengas, R.G.; Silva, V.L.M. Therapeutic Potential of Glycosyl Flavonoids as Anti-Coronaviral Agents. Pharmaceuticals 2021, 14, 546. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060546
Godinho PIC, Soengas RG, Silva VLM. Therapeutic Potential of Glycosyl Flavonoids as Anti-Coronaviral Agents. Pharmaceuticals. 2021; 14(6):546. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060546
Chicago/Turabian StyleGodinho, Patrícia I. C., Raquel G. Soengas, and Vera L. M. Silva. 2021. "Therapeutic Potential of Glycosyl Flavonoids as Anti-Coronaviral Agents" Pharmaceuticals 14, no. 6: 546. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060546