Genetic Markers for Coronary Artery Disease
Abstract
:1. Introduction
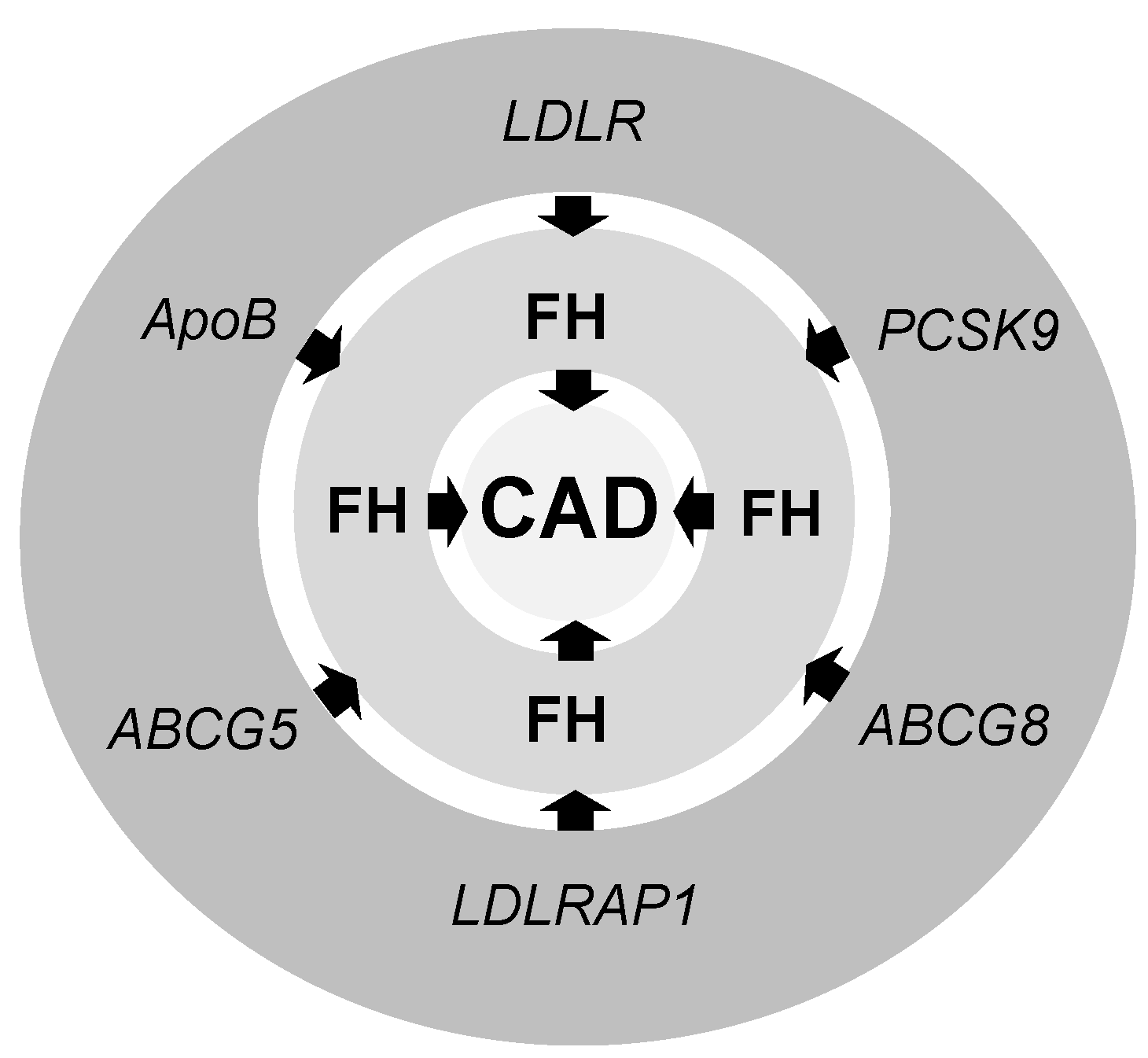
2. Familial Hypercholesterolemia (FH)
3. Genetic Markers for CAD
4. Low-Density Lipoprotein Receptor (LDLR)
5. Apolipoprotein B (ApoB)
6. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9)
7. LDLR Adaptor Protein 1 (LDLRAP1)
8. Adiponectin
9. C-Reactive Protein (CRP)
10. Ion Channels
11. GWAS Analysis and CAD
12. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Marenberg, M.E.; Risch, N.; Berkman, L.F.; Floderus, B.; De Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994, 330, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Zdravkovic, S.; Wienke, A.; Pedersen, N.L.; Marenberg, M.E.; Yashin, A.I.; De Faire, U. Heritability of death from coronary heart disease: A 36-year follow-up of 20 966 Swedish twins. J. Intern. Med. 2002, 252, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Catapano, A.L.; Lautsch, D.; Tokgozoglu, L.; Ferrieres, J.; Horack, M.; Farnier, M.; Toth, P.P.; Brudi, P.; Tomassini, J.E.; Ambegaonkar, B.; et al. Prevalence of potential familial hypercholesteremia (FH) in 54,811 statin-treated patients in clinical practice. Atherosclerosis 2016, 252, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, Y. Emerging Therapeutic Potential Targeting Genetics and Epigentics in Heart Failure. Biochim. Biophys. Acta 2017, 1863, 1867–1869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, J. Epigenetics and obesity cardiomyopathy: From pathophysiology to prevention and management. Pharmacol. Ther. 2016, 161, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Reiner, Z. Management of patients with familial hypercholesterolaemia. Nat. Rev. Cardiol. 2015, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.A.; Hutter, C.M.; Zimmern, R.L.; Humphries, S.E. Genetic causes of monogenic heterozygous familial hypercholesterolemia: A HuGE prevalence review. Am. J. Epidemiol. 2004, 160, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Paynter, N.P.; Ridker, P.M.; Chasman, D.I. Are Genetic Tests for Atherosclerosis Ready for Routine Clinical Use? Circ. Res. 2016, 118, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Risk of fatal coronary heart disease in familial hypercholesterolaemia. Scientific Steering Committee on behalf of the Simon Broome Register Group. BMJ 1991, 303, 893–896. [Google Scholar]
- Nordestgaard, B.G.; Chapman, M.J.; Humphries, S.E.; Ginsberg, H.N.; Masana, L.; Descamps, O.S.; Wiklund, O.; Hegele, R.A.; Raal, F.J.; Defesche, J.C.; et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: Guidance for clinicians to prevent coronary heart disease: Consensus statement of the European Atherosclerosis Society. Eur. Heart J. 2013, 34, 15. [Google Scholar] [CrossRef] [PubMed]
- Gidding, S.S.; Champagne, M.A.; De Ferranti, S.D.; Defesche, J.; Ito, M.K.; Knowles, J.W.; Mccrindle, B.; Raal, F.; Rader, D.; Santos, R.D.; et al. The Agenda for Familial Hypercholesterolemia: A Scientific Statement from the American Heart Association. Circulation 2015, 132, 2167–2192. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; O’kane, M.; Mcgilligan, V.; Watterson, S. The genetics and screening of familial hypercholesterolaemia. J. Biomed. Sci. 2016, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Rabes, J.; Varret, M.; Devillers, M.; Aegerter, P.; Villéger, L.; Krempf, M.; Junien, C.; Boileau, C.; Rabes, J.P.; Varret, M.; et al. R3531C mutation in the apolipoprotein B gene is not sufficient to cause hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2000, 20, E76–E82. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Won, H.H.; Peloso, G.M.; Lawson, K.S.; Bartz, T.M.; Deng, X.; Van Leeuwen, E.M.; Natarajan, P.; Emdin, C.A.; Bick, A.G.; et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients with Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016, 67, 2578–2589. [Google Scholar] [CrossRef] [PubMed]
- Sijbrands, E.J.G.; Westendorp, R.G.J.; Defesche, J.C.; De Meier, P.H.E.M.; Smelt, A.H.M.; Kastelein, J.J.P. Mortality over two centuries in large pedigree with familial hypercholesterolaemia: Family tree mortality study. BMJ Br. Med. J. 2001, 322, 1019–1023. [Google Scholar] [CrossRef]
- Hill, J.; Hayden, M.; Frohlich, J.; Pritchard, P. Genetic and environmental factors affecting the incidence of coronary artery disease in heterozygous familial hypercholesterolemia. Arterioscler. Thromb. 1991, 11, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, P.J.; Sukumar, A.N.; Marsden, P.A. Epigenetics of Cardiovascular Disease-A New “Beat” in Coronary Artery Disease. Med. Epigenet. 2014, 2, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Kathiresan, S. Genetics of coronary artery disease. Annu. Rev. Genom. Hum. Genet. 2010, 11, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhao, S. Candidate gene identification approach: Progress and challenges. Int. J. Biol. Sci. 2007, 3, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Kohli, P.; Baber, U.; Nguyen, K.H.; Sartori, S.; Reilly, D.F.; Mehran, R.; Muntendam, P.; Fuster, V.; Rader, D.J.; et al. Association of APOC3 Loss-of-Function Mutations with Plasma Lipids and Subclinical Atherosclerosis: The Multi-Ethnic BioImage Study. J. Am. Coll. Cardiol. 2015, 66, 2053–2055. [Google Scholar] [CrossRef] [PubMed]
- Glisic, S.; Arrigo, P.; Alavantic, D.; Perovic, V.; Prljic, J.; Veljkovic, N. Lipoprotein lipase: A bioinformatics criterion for assessment of mutations as a risk factor for cardiovascular disease. Proteins 2008, 70, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, A.; Willems, S.M.; Bos, D.; Dehghan, A.; Hofman, A.; Ikram, M.A.; Uitterlinden, A.G.; Oostra, B.A.; Franco, O.H.; Witteman, J.C.; et al. Risk scores of common genetic variants for lipid levels influence atherosclerosis and incident coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Embl-Eb. The NHGRI-EBI Catalog of Published Genome-Wide Association Studies. 2018. Available online: https://www.ebi.ac.uk/gwas/home (accessed on 23 June 2017).
- Stranger, B.E.; Stahl, E.A.; Raj, T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 2011, 187, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Abecasis, G.R.; Boehnke, M.; Lin, X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014, 95, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Toth, P.P. The Year in Lipid Disorders, 1st ed.; Oxford Centre for Innovation Mill Street: Oxford, UK, 2010. [Google Scholar]
- Beglova, N.; Jeon, H.; Fisher, C.; Blacklow, S.C. Structural features of the low-density lipoprotein receptor facilitating ligand binding and release. Biochem. Soc. Trans. 2004, 32, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, G.; Henry, L.; Henderson, K.; Ichtchenko, K.; Brown, M.S.; Goldstein, J.L.; Deisenhofer, J. Structure of the LDL receptor extracellular domain at endosomal pH. Science 2002, 298, 2353–2358. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem. Sci. 2007, 32, 71–77. [Google Scholar] [CrossRef] [PubMed]
- North, C.L.; Blacklow, S.C. Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry 2000, 39, 2564–2571. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.D. Familial Hypercholesterolaemia. Clin. Biochem. Rev. 2004, 25, 49–68. [Google Scholar] [PubMed]
- Jones, C.; Hammer, R.E.; Li, W.P.; Cohen, J.C.; Hobbs, H.H.; Herz, J. Normal sorting but defective endocytosis of the low density lipoprotein receptor in mice with autosomal recessive hypercholesterolemia. J. Biol. Chem. 2003, 278, 29024–29030. [Google Scholar] [CrossRef] [PubMed]
- Farnier, M. The role of proprotein convertase subtilisin/kexin type 9 in hyperlipidemia: Focus on therapeutic implications. Am. J. Cardiovasc. Drugs 2011, 11, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Contois, J.H.; Mcconnell, J.P.; Sethi, A.A.; Csako, G.; Devaraj, S.; Hoefner, D.M.; Warnick, G.R.; Lipoproteins, A. Vascular Diseases Division Working Group on Best P. Apolipoprotein B and cardiovascular disease risk: Position statement from the AACC Lipoproteins and Vascular Diseases Division Working Group on Best Practices. Clin. Chem. 2009, 55, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Kaneva, A.M.; Potolitsyna, N.N.; Bojko, E.R.; Odland, J.O. The apolipoprotein B/apolipoprotein A–I ratio as a potential marker of plasma atherogenicity. Dis. Markers 2015, 2015, 591454. [Google Scholar] [CrossRef] [PubMed]
- Sniderman, A.D.; Pedersen, T.; Kjekshus, J. Putting low-density lipoproteins at center stage in atherogenesis. Am. J. Cardiol. 1997, 79, 64–67. [Google Scholar] [PubMed]
- Sniderman, A.; Vu, H.; Cianflone, K. Effect of moderate hypertriglyceridemia on the relation of plasma total and LDL apo B levels. Atherosclerosis 1991, 89, 109–116. [Google Scholar] [CrossRef]
- Vakkilainen, J.; Steiner, G.; Ansquer, J.C.; Aubin, F.; Rattier, S.; Foucher, C.; Hamsten, A.; Taskinen, M.R. Relationships between low-density lipoprotein particle size, plasma lipoproteins, and progression of coronary artery disease: The Diabetes Atherosclerosis Intervention Study (DAIS). Circulation 2003, 107, 1733–1737. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, M.; Pernice, V.; Frasheri, A.; Berneis, K. Atherogenic lipoprotein phenotype and LDL size and subclasses in patients with peripheral arterial disease. Atherosclerosis 2008, 197, 237–241. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, A.C.; Ruel, I.L.; Cantin, B.; Dagenais, G.R.; Bernard, P.M.; Després, J.P.; Lamarche, B. Comparison of Various Electrophoretic Characteristics of LDL Particles and Their Relationship to the Risk of Ischemic Heart Disease. Circulation 2001, 104, 2295–2299. [Google Scholar] [CrossRef] [PubMed]
- Zambon, A.; Hokanson, J.E.; Brown, B.G.; Brunzell, J.D. Evidence for a New Pathophysiological Mechanism for Coronary Artery Disease Regression. Circulation 1999, 99, 1959–1964. [Google Scholar] [CrossRef] [PubMed]
- Young, S.G. Recent progress in understanding apolipoprotein, B. Circulation 1990, 82, 1574–1594. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.M.; Nordskog, B.K.; Nauli, A.M.; Zheng, S.; Vonlehmden, S.B.; Yang, Q.; Lee, D.; Swift, L.L.; Davidson, N.O.; Tso, P. Why does the gut choose apolipoprotein B48 but not B100 for chylomicron formation? Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G344–G352. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Li, X.X.; Liao, W.S.; Wu, J.H.; Chan, L. RNA editing of apolipoprotein B mRNA. Sequence specificity determined by in vitro coupled transcription editing. J. Biol. Chem. 1990, 265, 6811–6816. [Google Scholar] [PubMed]
- Nakamuta, M.; Oka, K.; Krushkal, J.; Kobayashi, K.; Yamamoto, M.; Li, W.H.; Chan, L. Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice. Structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases. J. Biol. Chem. 1995, 270, 13042–13056. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. A century of cholesterol and coronaries: From plaques to genes to statins. Cell 2015, 161, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Banach, M.; Rizzo, M.; Obradovic, M.; Montalto, G.; Rysz, J.; Mikhailidis, D.P.; Isenovic, E.R. PCSK9 inhibition-a novel mechanism to treat lipid disorders? Curr. Pharm. Des. 2013, 19, 3869–3877. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, M.; Zaric, B.; Sudar-Milovanovic, E.; Ilincic, B.; Perovic, M.; Stokic, E.; Isenovic, E. PCSK9 and hypercholesterolemia: Therapeutical approach. Curr. Drug Targets 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Katsiki, N.; Giannoukas, A.D.; Athyros, V.G.; Mikhailidis, D.P. Lipid-lowering treatment in peripheral artery disease. Curr. Opin. Pharmacol. 2018, 39, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Chretien, M. Proprotein and prohormone convertases of the subtilisin family Recent developments and future perspectives. Trends Endocrinol. MeTable 1992, 3, 133–140. [Google Scholar] [CrossRef]
- Zhang, L.; Song, K.; Zhu, M.; Shi, J.; Zhang, H.; Xu, L.; Chen, Y. Proprotein convertase subtilisin/kexin type 9 (PCSK9) in lipid metabolism, atherosclerosis and ischemic stroke. Int. J. Neurosci. 2016, 126, 675–680. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, P.; Elbitar, S.; Ghaleb, Y.; Khalil, Y.A.; Varret, M.; Boileau, C.; Abifadel, M. PCSK9 Mutations in Familial Hypercholesterolemia: From a Groundbreaking Discovery to Anti-PCSK9 Therapies. Curr. Atheroscler. Rep. 2017, 19, 49. [Google Scholar] [CrossRef] [PubMed]
- Basic Local Alignment Search Tool. Available online: www.ncbi.nlm.nih.gov/BLAST (accessed on 23 June 2017).
- Seidah, N.G.; Benjannet, S.; Wickham, L.; Marcinkiewicz, J.; Jasmin, S.B.; Stifani, S.; Basak, A.; Prat, A.; Chretien, M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): Liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA 2003, 100, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Abifadel, M.; Varret, M.; Rabes, J.P.; Allard, D.; Ouguerram, K.; Devillers, M.; Cruaud, C.; Benjannet, S.; Wickham, L.; Erlich, D.; et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003, 34, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Nissen, S.E.; Nicholls, S.J.; Sipahi, I.; Libby, P.; Raichlen, J.S.; Ballantyne, C.M.; Davignon, J.; Erbel, R.; Fruchart, J.C.; Tardif, J.C.; et al. Effect of very high-intensity statin therapy on regression of coronary atherosclerosis: The ASTEROID trial. JAMA 2006, 295, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. Molecular medicine. The cholesterol quartet. Science 2001, 292, 1310–1312. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Soccio, R.E.; Duncan, E.M.; Sehayek, E.; Breslow, J.L. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 2003, 44, 2109–2119. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.K.; Wilund, K.; Arca, M.; Zuliani, G.; Fellin, R.; Maioli, M.; Calandra, S.; Bertolini, S.; Cossu, F.; Grishin, N.; et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 2001, 292, 1394–1398. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Watkins, S.C.; Traub, L.M. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl. Acad. Sci. USA 2002, 99, 16099–16104. [Google Scholar] [CrossRef] [PubMed]
- Usifo, E.; Leigh, S.E.; Whittall, R.A.; Lench, N.; Taylor, A.; Yeats, C.; Orengo, C.A.; Martin, A.C.; Celli, J.; Humphries, S.E. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: Update and pathological assessment. Ann. Hum. Genet. 2012, 76, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Ryo, M.; Nakamura, T.; Kihara, S.; Kumada, M.; Shibazaki, S.; Takahashi, M.; Nagai, M.; Matsuzawa, Y.; Funahashi, T. Adiponectin as a biomarker of the metabolic syndrome. Circ. J. 2004, 68, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Lihn, A.S.; Pedersen, S.B.; Richelsen, B. Adiponectin: Action, regulation and association to insulin sensitivity. Obes. Rev. 2005, 6, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, A.; Yan, Y.; Song, F.; Kong, Q.; Qin, S.; Zhang, D. Association between HMW adiponectin, HMW-total adiponectin ratio and early-onset coronary artery disease in Chinese population. Atherosclerosis 2014, 235, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, R.S.; Funahashi, T.; Hanson, R.L.; Matsuzawa, Y.; Tanaka, S.; Tataranni, P.A.; Knowler, W.C.; Krakoff, J. Adiponectin and development of type 2 diabetes in the Pima Indian population. Lancet 2002, 360, 57–58. [Google Scholar] [CrossRef]
- Nakamura, Y.; Shimada, K.; Fukuda, D.; Shimada, Y.; Ehara, S.; Hirose, M.; Kataoka, T.; Kamimori, K.; Shimodozono, S.; Kobayashi, Y.; et al. Implications of plasma concentrations of adiponectin in patients with coronary artery disease. Heart 2004, 90, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Spranger, J.; Kroke, A.; Mohlig, M.; Bergmann, M.M.; Ristow, M.; Boeing, H.; Pfeiffer, A.F. Adiponectin and protection against type 2 diabetes mellitus. Lancet 2003, 361, 226–228. [Google Scholar] [CrossRef]
- Hotta, K.; Funahashi, T.; Arita, Y.; Takahashi, M.; Matsuda, M.; Okamoto, Y.; Iwahashi, H.; Kuriyama, H.; Ouchi, N.; Maeda, K.; et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1595–1599. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.S.; Tayebjee, M.H.; Tan, K.T.; Patel, J.V.; Macfadyen, R.J.; Lip, G.Y. Serum adiponectin in coronary heart disease: Ethnic differences and relation to coronary artery disease severity. Heart 2005, 91, 1605–1606. [Google Scholar] [CrossRef] [PubMed]
- Khan, U.I.; Wang, D.; Sowers, M.R.; Mancuso, P.; Everson-Rose, S.A.; Scherer, P.E.; Wildman, R.P. Race-ethnic differences in adipokine levels: The Study of Women’s Health Across the Nation (SWAN). Metabolism 2012, 61, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Hulver, M.W.; Saleh, O.; Macdonald, K.G.; Pories, W.J.; Barakat, H.A. Ethnic differences in adiponectin levels. Metabolism 2004, 53, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Torzewski, M.; Rist, C.; Mortensen, R.F.; Zwaka, T.P.; Bienek, M.; Waltenberger, J.; Koenig, W.; Schmitz, G.; Hombach, V.; Torzewski, J. C-reactive protein in the arterial intima: Role of C-reactive protein receptor-dependent monocyte recruitment in atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 2094–2099. [Google Scholar] [CrossRef] [PubMed]
- Zakynthinos, E.; Pappa, N. Inflammatory biomarkers in coronary artery disease. J. Cardiol. 2009, 53, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Mach, F.; Lovis, C.; Gaspoz, J.M.; Unger, P.F.; Bouillie, M.; Urban, P.; Rutishauser, W. C-reactive protein as a marker for acute coronary syndromes. Eur. Heart J. 1997, 18, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Kaptoge, S.; Di Angelantonio, E.; Lowe, G.; Pepys, M.B.; Thompson, S.G.; Collins, R.; Danesh, J. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: An individual participant meta-analysis. Lancet 2010, 375, 132–140. [Google Scholar] [PubMed]
- Van Wijk, D.F.; Boekholdt, S.M.; Wareham, N.J.; Ahmadi-Abhari, S.; Kastelein, J.J.; Stroes, E.S.; Khaw, K.T. C-reactive protein, fatal and nonfatal coronary artery disease, stroke, and peripheral artery disease in the prospective EPIC-Norfolk cohort study. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2888–2894. [Google Scholar] [CrossRef] [PubMed]
- Auer, J.; Berent, R.; Lassnig, E.; Eber, B. C-reactive protein and coronary artery disease. Jpn. Heart J. 2002, 43, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.S.; Al Masri, A.A. Relationship of high sensitivity C-reactive protein with presence and severity of coronary artery disease. Pak. J. Med. Sci. 2013, 29, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Ockene, I.S.; Matthews, C.E.; Rifai, N.; Ridker, P.M.; Reed, G.; Stanek, E. Variability and classification accuracy of serial high-sensitivity C-reactive protein measurements in healthy adults. Clin. Chem. 2001, 47, 444–450. [Google Scholar] [PubMed]
- Fedele, F.; Mancone, M.; Chilian, W.M.; Severino, P.; Canali, E.; Logan, S.; De Marchis, M.L.; Volterrani, M.; Palmirotta, R.; Guadagni, F. Role of genetic polymorphisms of ion channels in the pathophysiology of coronary microvascular dysfunction and ischemic heart disease. Basic Res. Cardiol. 2013, 108, 387. [Google Scholar] [CrossRef] [PubMed]
- Fedele, F.; Severino, P.; Bruno, N.; Stio, R.; Caira, C.; D’ambrosi, A.; Brasolin, B.; Ohanyan, V.; Mancone, M. Role of ion channels in coronary microcirculation: A review of the literature. Future Cardiol. 2013, 9, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; D’amato, A.; Netti, L.; Pucci, M.; De Marchis, M.; Palmirotta, R.; Volterrani, M.; Mancone, M.; Fedele, F. Diabetes Mellitus and Ischemic Heart Disease: The Role of Ion Channels. Int. J. Mol. Sci. 2018, 19, 802. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Parast, L.; Cai, T.; Powers, C.; Gervino, E.V.; Hauser, T.H.; Hu, F.B.; Doria, A. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J. Am. Coll. Cardiol. 2011, 58, 2675–2682. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.D.; Manichaikul, A.; Wang, X.Q.; Rich, S.S.; Rotter, J.I.; Post, W.S.; Polak, J.F.; Budoff, M.J.; Bluemke, D.A. Common genetic variants and subclinical atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis 2016, 245, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Campillo, A.; Roberts, R. Discovery of six new genetic risk variants predisposing to CAD. Cardiology Today, June 2017. Available online: https://0-www-healio-com.brum.beds.ac.uk/cardiology/genetics-genomics/news/print/cardiology-today/%7B26cd2dbc-9e44-4c5e-9c28-e35daa8011c3%7D/discovery-of-six-new-genetic-risk-variants-predisposing-to-cad (accessed on 23 June 2017).
- Barth, A.S.; Tomaselli, G.F. Gene scanning and heart attack risk. Trends Cardiovasc. Med. 2016, 26, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R. A Breakthrough in Genetics and its Relevance to Prevention of Coronary Artery Disease in LMIC. Glob. Heart 2017, 12, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Schunkert, H.; Erdmann, J.; Samani, N.J. Genetics of myocardial infarction: A progress report. Eur. Heart J. 2010, 31, 918–925. [Google Scholar] [CrossRef] [PubMed]
- So, H.C.; Gui, A.H.; Cherny, S.S.; Sham, P.C. Evaluating the heritability explained by known susceptibility variants: A survey of ten complex diseases. Genet. Epidemiol. 2011, 35, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Mcpherson, R.; Pertsemlidis, A.; Kavaslar, N.; Stewart, A.; Roberts, R.; Cox, D.R.; Hinds, D.A.; Pennacchio, L.A.; Tybjaerg-Hansen, A.; Folsom, A.R.; et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007, 316, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; Masson, G.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Magnusson, K.P.; Gretarsdottir, S.; Steinthorsdottir, V.; Manolescu, A.; Jones, G.T.; Rinkel, G.J.; Blankensteijn, J.D.; Ronkainen, A.; et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat. Genet. 2008, 40, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Lanktree, M.B.; Hegele, R.A. Gene-gene and gene-environment interactions: New insights into the prevention, detection and management of coronary artery disease. Genome Med. 2009, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Hartiala, J.; Schwartzman, W.S.; Gabbay, J.; Ghazalpour, A.; Bennett, B.J.; Allayee, H. The Genetic Architecture of Coronary Artery Disease: Current Knowledge and Future Opportunities. Curr. Atheroscler. Rep. 2017, 19, 6. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.B.; Nikpay, M.; Lau, P.; Stewart, A.F.; Davies, R.W.; Wells, G.A.; Dent, R.; Mcpherson, R. Adiposity significantly modifies genetic risk for dyslipidemia. J. Lipid Res. 2014, 55, 2416–2422. [Google Scholar] [CrossRef] [PubMed]
- Euesden, J.; Lewis, C.M.; O’reilly, P.F. PRSice: Polygenic Risk Score software. Bioinformatics 2015, 31, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013, 9, e1003348. [Google Scholar] [CrossRef]
- Mega, J.L.; Stitziel, N.O.; Smith, J.G.; Chasman, D.I.; Caulfield, M.; Devlin, J.J.; Nordio, F.; Hyde, C.; Cannon, C.P.; Sacks, F.; et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: An analysis of primary and secondary prevention trials. Lancet 2015, 385, 2264–2271. [Google Scholar] [CrossRef]
- Tada, H.; Melander, O.; Louie, J.Z.; Catanese, J.J.; Rowland, C.M.; Devlin, J.J.; Kathiresan, S.; Shiffman, D. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur. Heart J. 2016, 37, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Ganna, A.; Magnusson, P.K.; Pedersen, N.L.; De Faire, U.; Reilly, M.; Arnlov, J.; Sundstrom, J.; Hamsten, A.; Ingelsson, E. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2267–2272. [Google Scholar] [CrossRef] [PubMed]
- Ripatti, S.; Tikkanen, E.; Orho-Melander, M.; Havulinna, A.S.; Silander, K.; Sharma, A.; Guiducci, C.; Perola, M.; Jula, A.; Sinisalo, J.; et al. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet 2010, 376, 1393–1400. [Google Scholar] [CrossRef]
- Thanassoulis, G.; Vasan, R.S. Genetic cardiovascular risk prediction: Will we get there? Circulation 2010, 122, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.W. Challenges to improve coronary heart disease risk assessment. JAMA 2009, 302, 2369–2370. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veljkovic, N.; Zaric, B.; Djuric, I.; Obradovic, M.; Sudar-Milovanovic, E.; Radak, D.; Isenovic, E.R. Genetic Markers for Coronary Artery Disease. Medicina 2018, 54, 36. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54030036
Veljkovic N, Zaric B, Djuric I, Obradovic M, Sudar-Milovanovic E, Radak D, Isenovic ER. Genetic Markers for Coronary Artery Disease. Medicina. 2018; 54(3):36. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54030036
Chicago/Turabian StyleVeljkovic, Nevena, Bozidarka Zaric, Ilona Djuric, Milan Obradovic, Emina Sudar-Milovanovic, Djordje Radak, and Esma R. Isenovic. 2018. "Genetic Markers for Coronary Artery Disease" Medicina 54, no. 3: 36. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54030036




