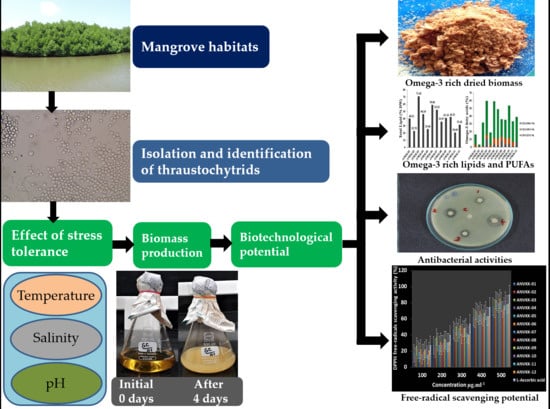
Thraustochytrids of Mangrove Habitats from Andaman Islands: Species Diversity, PUFA Profiles and Biotechnological Potential
Abstract
:1. Introduction
2. Results and Discussion
2.1. Morphological and Molecular Identification
2.2. Effect of Temperature, Salinity, and pH Tolerance
2.3. Growth Behavior of Thraustochytrids
2.4. Biomass Production of Thraustochytrids
2.5. Lipid and Fatty Acid Production
2.6. Blood Agar Hemolysis Activity
2.7. Antibacterial Activity
2.8. Antioxidant Activity
3. Material and Methods
3.1. Sample Collection and Processing
3.2. Isolation and Maintenance of Thraustochytrids
3.3. Morphological Identification of Thraustochytrids by Light Microscope (LM) and Scanning Electron Microscope (SEM)
3.4. Nile Red Staining
3.5. Genomic DNA Extraction, Sequencing, and Phylogenetic Analysis
3.6. Effect of Temperature, Salinity, and pH Tolerance
3.7. Observation of Thraustochytrids Growth Behavior
3.8. Biomass Production of Thraustochytrids
3.9. Lipid Extraction and Fatty Acid Methyl Ester Analysis
3.10. Blood Agar Hemolysis Activity
3.11. Screening of Antibacterial Activity
3.12. Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bongiorni, L. Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 53, ISBN 3642233414. [Google Scholar]
- Armenta, R.E.; Valentine, M.C. Single-cell oils as a source of omega-3 fatty acids: An overview of recent advances. J. Am. Oil Chem. Soc. 2012, 90, 167–182. [Google Scholar] [CrossRef]
- Leyland, B.; Leu, S.; Boussibax, S. Are thraustochytrids algae? Fungal. Biol. 2017, 121, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Aki, T.; Yokochi, T.; Nakahara, T.; Honda, D.; Kawamoto, S.; Shigeta, S.; Ono, K.; Suzuki, O. Grouping newly isolated docosahexaenoic acid-producing thraustochytrids based on their polyunsaturated fatty acid profiles and comparative analysis of 18S rRNA genes. Mar. Biotechnol. 2003, 5, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Bongiorni, L.; Jain, R.; Raghukumar, S.; Aggarwal, R. Thraustochytrium gaertnerium sp. nov.: A new thraustochytrid stramenopilan protist from mangroves of Goa, India. Protist 2005, 156, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Honda, D. Taxonomic rearrangement of the genus Schizochytrium sensu lato based on morphology, chemotaxonomic characteristics, and 18S rRNA gene phylogeny (Thraustochytriaceae, Labyrinthulomycetes): Emendation for Schizochytrium and erection of Aurantiochytrium and Oblongichytrium gen. nov. Mycoscience 2007, 48, 199–211. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Alderman, D.J. Althornia crouchii gen. et sp. nov. A marine biflagellate fungus. Nova Hedwigia 1971, 21, 381–399. [Google Scholar]
- FioRito, R.; Leander, C.; Leander, B. Characterization of three novel species of Labyrinthulomycota isolated from ochre sea stars (Pisaster ochraceus). Mar. Biol. 2016, 163, 1–10. [Google Scholar] [CrossRef]
- Sparrow, F.K. Biological observations on the marine fungi of woods hole waters. Biol. Bull. 1936, 70, 236–263. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ookubo, M. Studies on the marine Phycomycetes. Bull. Natl. Sci. Mus. Tokyo 1953, 33, 53–65. [Google Scholar]
- Goldstein, S.; Belsky, M. Axenic culture studies of a new marine phycomycete possessing an unusual type of asexual reproduction. Am. J. Bot. 1964, 51, 72–78. [Google Scholar] [CrossRef]
- Gaertner, A. Revision of the Thraustochytriaceae (lower marine fungi)—I. Ulkenia nov. gen., with description of three new species. Veroff. Inst. Meeresforsch. Bremerh. 1977, 16, 139–157. [Google Scholar] [CrossRef]
- Yokoyama, R.; Salleh, B.; Honda, D. Taxonomic rearrangement of the genus Ulkenia sensu lato based on morphology, chemotaxonomical characteristics, and 18S rRNA gene phylogeny (Thraustochytriaceae, Labyrinthulomycetes): Emendation for Ulkenia and erection of Botryochytrium, Parietichytrium, and Sicyoidochytrium gen. nov. Mycoscience 2007, 48, 329–341. [Google Scholar] [CrossRef]
- Doi, K.; Honda, D. Proposal of Monorhizochytrium globosum gen. nov., comb. nov. (Stramenopiles, Labyrinthulomycetes) for former Thraustochytrium globosum based on morphological features and phylogenetic relationships. Phycol. Res. 2017, 65, 188–201. [Google Scholar] [CrossRef]
- Dellero, Y.; Cagnac, O.; Rose, S.; Seddiki, K.; Cussac, M.; Morabito, C.; Lupette, J.; Cigliano, R.A.; Sanseverino, W.; Kuntz, M.; et al. Proposal of a new thraustochytrid genus Hondaea gen. nov. and comparison of its lipid dynamics with the closely related pseudo-cryptic genus Aurantiochytrium. Algal Res. 2018, 35, 125–141. [Google Scholar] [CrossRef]
- Hassett, B.T.; Gradinger, R. New Species of Saprobic Labyrinthulea (=Labyrinthulomycota) and the Erection of a gen. nov. to resolve molecular polyphyly within the Aplanochytrids. J. Eukaryot. Microbiol. 2018, 65, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghukumar, S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistol. 2002, 38, 127–145. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, H.; Xie, Y.; He, Y.; Sen, B.; Wang, G. Culturable diversity and lipid production profile of Labyrinthulomycete protists isolated from coastal mangrove habitats of China. Mar. Drugs 2019, 17, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalidasan, K.; Phusit, H.; Kathiresan, K. Enumeration of thraustochytrids in decomposing leaves of mangroves as influenced by physicochemical and microbial factors. J. Curr. Rese. Env. App. Myco. 2019, 9, 288–300. [Google Scholar] [CrossRef]
- Taoka, Y.; Nagano, N.; Okita, Y.; Izumida, H.; Sugimoto, S.; Hayashi, M. Extracellular enzymes produced by marine eukaryotes, thraustochytrids. Biosci. Biotechnol. Biochem. 2009, 73, 180–182. [Google Scholar] [CrossRef] [Green Version]
- Taoka, Y.; Nagano, N.; Kai, H.; Hayashi, M. Degradation of distillery lees (Shochu kasu) by cellulase-producing thraustochytrids. J. Oleo Sci. 2017, 66, 31–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalidasan, K.; Asmathunisha, N.; Gomathi, V.; Dufossé, L.; Kathiresan, K. Isolation and optimization of culture conditions of Thraustochytrium kinnei for biomass production, nanoparticle synthesis, antioxidant and antimicrobial activities. J. Mar. Sci. Eng. 2021, 9, 678. [Google Scholar] [CrossRef]
- Raghukumar, S. Thraustochytrid marine protists: Production of PUFAs and other emerging technologies. Mar. Biotechnol. 2008, 10, 631–640. [Google Scholar] [CrossRef]
- Sprague, M.; Walton, J.; Campbell, P.; Strachan, F.; Dick, J.; Bell, J. Replacement of fish oil with a DHA-rich algal meal derived from Schizochytrium sp. on the fatty acid and persistent organic pollutant levels in diets and flesh of Atlantic salmon (Salmo salar, L.) post-smolts. Food Chem. 2015, 185, 413–421. [Google Scholar] [CrossRef]
- Polyunsaturated fatty acid-producing marine thraustochytrids: A potential source for antimicrobials. J. Coast. Life Med. 2015, 3, 848–851. [CrossRef]
- Kalidasan, K.; Sunil, K.S.; Narendran, R.; Kathiresan, K. Antioxidant activity of mangrove-derived marine thraustochytrids. Mycosphere 2015, 6, 602–611. [Google Scholar] [CrossRef]
- Simopoulos, A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1999, 70, 560s–569s. [Google Scholar] [CrossRef] [Green Version]
- Shakeri, S.; Amoozyan, N.; Fekrat, F.; Maleki, M. Antigastric cancer bioactive Aurantiochytrium oil rich in docosahexaenoic acid: From media optimization to cancer cells cytotoxicity assessment. J. Food Sci. 2017, 82, 2706–2718. [Google Scholar] [CrossRef]
- Innis, S.M. Dietary omega 3 fatty acids and the developing brain. Brain Res. 2008, 1237, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kathiresan, K.; Qasim, S.Z. Biodiversity of Mangrove Ecosystems; Hindustan Publication Corporation: New Delhi, India, 2005; p. 251. ISBN 81-7075-079-2. [Google Scholar]
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 81–251. [Google Scholar] [CrossRef]
- Ragavan, P.; Saxena, A.; Jayaraj, R.S.C.; Mohan, P.M.; Ravichandran, K.; Saravanan, S.; Vijayaraghavan, A. A review of the mangrove floristic of India. Taiwania 2016, 61, 224–242. [Google Scholar] [CrossRef]
- Fan, K.W.; Vrijmoed, L.L.P.; Jones, E.B.G. Physiological studies of subtropical mangrove thraustochytrids. Bot. Mar. 2002, 45, 50–57. [Google Scholar] [CrossRef]
- Raghukumar, S.; Sathe-Pathak, V.; Sharma, S.; Raghukumar, C. Thraustochytrid and fungal component of marine detritus. III. Field studies on decomposition of leaves of the mangrove Rhizophora apiculata. Aquat. Microb. Ecol. 1995, 9, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Jaseera, K.V.; Kaladharan, P.; Vijayan, K.K.; Sandhya, S.V.; Antony, M.L.; Pradeep, M.A. Isolation and phylogenetic identification of heterotrophic thraustochytrids from mangrove habitats along the southwest coast of India and prospecting their PUFA accumulation. Environ. Boil. Fishes 2018, 31, 1057–1068. [Google Scholar] [CrossRef]
- Pandey, A.; Bhathena, Z. Prevalence of PUFA rich Thraustochytrids sps. along the coast of mumbai for production of bio oil. J. Food Nutr. Res. 2014, 2, 993–999. [Google Scholar] [CrossRef] [Green Version]
- Bremer, G. Isolation and Culture of Thraustochytrids. Marine Mycology—A Practical Approach; Fungal Diversity Press: Hong Kong, China, 2000; pp. 49–61. [Google Scholar]
- Kimura, H.; Fukuba, T.; Naganuma, T. Biomass of thraustochytrid protoctists in coastal water. Mar. Ecol. Prog. Ser. 1999, 189, 27–33. [Google Scholar] [CrossRef]
- Raghukumar, S.; Schaumann, K. An epifluorescence microscopy method for direct detection and enumeration of the fungi like marine protists, the thraustochytrids. Limnol. Oceanogra 1993, 38, 182–187. [Google Scholar]
- Marchan, L.F.; Chang, K.J.L.; Nichols, P.D.; Mitchell, W.J.; Polglase, J.; Gutierrez, T. Taxonomy, ecology and biotechnological applications of thraustochytrids: A review. Biotechnol. Adv. 2018, 36, 26–46. [Google Scholar] [CrossRef]
- Dick, M.W. Straminipilous Fungi. Systematics of the Peronosporomycetes Including Accounts of the Marine Straminipilous Protists, the Plasmodiophorids and Similar Organisms; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001. [Google Scholar]
- Leander, C.A.; Porter, D.; Leander, B.S. Comparative morphology and molecular phylogeny of aplanochytrids (Labyrinthulomycota). Eur. J. Protistol. 2004, 40, 317–328. [Google Scholar] [CrossRef]
- Unagul, P.; Suetrong, S.; Preedanon, S.; Klaysuban, A.; Gundool, W.; Suriyachadkun, C.; Sakayaroj, J. Isolation, fatty acid profiles and cryopreservation of marine thraustochytrids from mangrove habitats in Thailand. Bot. Mar. 2017, 60, 363–379. [Google Scholar] [CrossRef]
- Kumon, Y.; Yokoyama, R.; Haque, Z.; Yokochi, T.; Honda, D.; Nakahara, T. A New Labyrinthulid isolate that produces only docosahexaenoic acid. Mar. Biotechnol. 2006, 8, 170–177. [Google Scholar] [CrossRef]
- Perez-Garcia, O.; Escalante, F.M.E.; De-Bashan, L.E.; Bashan, Y. Heterotrophic cultures of microalgae: Metabolism and potential products. Water Res. 2011, 45, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Leano, E.M.; Gapasin, R.S.J.; Polohan, B.; Vrijmoed, L.L.P. Growth and fatty acid production of thraustochytrids from Panay mangroves, Philippines. Fungal Diversi. 2003, 12, 111–122. [Google Scholar]
- Caamaño, E.; Loperena, L.; Hinzpeter, I.; Pradel, P.; Gordillo, F.; Corsini, G.; Tello, M.; Lavín, P.; González, A.R. Isolation and molecular characterization of Thraustochytrium strain isolated from antarctic peninsula and its biotechnological potential in the production of fatty acids. Braz. J. Microbiol. 2017, 48, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, X.; Ren, X.; Zhu, Q. Effects of culture conditions on growth and docosahexaenoic acid production from Schizochytrium limacinum. J. Ocean Univ. China 2008, 7, 83–88. [Google Scholar] [CrossRef]
- Yaguchi, T.; Tanaka, S.; Yokochi, T.; Nakahara, T.; Higashihara, T. Production of high yields of docosahexaenoic acid by Schizochytrium sp. strain SR21. J. Am. Oil Chem. Soc. 1997, 74, 1431–1434. [Google Scholar] [CrossRef]
- Arafiles, K.H.V.; Alcantara, J.C.O.; Cordero, P.R.F.; Batoon, J.A.L.; Galura, F.S.; Leano, E.M.; Dedeles, G.R. Cultural optimization of thraustochytrids for biomass and fatty acid production. Mycosphere 2011, 2, 521–531. [Google Scholar]
- Kamlangdee, N.; Fan, K.W. Polyunsaturated fatty acids production by Schizochytrium sp. isolated from mangrove, Songklanakarin. J. Sci. Tech. 2003, 25, 643–650. [Google Scholar]
- Rau, E.-M.; Aasen, I.M.; Ertesvåg, H. A non-canonical Δ9-desaturase synthesizing palmitoleic acid identified in the thraustochytrid Aurantiochytrium sp. T66. Appl. Microbiol. Biotechnol. 2021, 105, 5931–5941. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Wilkens, S.; Adcock, J.L.; Puri, M.; Barrow, C.J. Pollen baiting facilitates the isolation of marine thraustochytrids with potential in omega-3 and biodiesel production. J. Ind. Microbiol. Biotechnol. 2013, 40, 1231–1240. [Google Scholar] [CrossRef]
- Chang, K.J.L.; Nichols, C.M.; Blackburn, S.I.; Dunstan, G.A.; Koutoulis, A.; Nichols, P.D. Comparison of Thraustochytrids Aurantiochytrium sp., Schizochytrium sp., Thraustochytrium sp., and Ulkenia sp. for production of biodiesel, long-chain omega-3 oils, and exopolysaccharide. Mar. Biotechnol. 2014, 16, 396–411. [Google Scholar] [CrossRef]
- Scott, S.D.; Armenta, R.E.; Berryman, K.T.; Norman, A.W. Use of raw glycerol to produce oil rich in polyunsaturated fatty acids by a thraustochytrid. Enzym. Microb. Technol. 2011, 48, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Yokochi, T.; Honda, D.; Higashihara, T.; Nakahara, T. Optimization of docosahexaenoic acid production by Schizochytrium limacinum SR21. Appl. Microbiol. Biotechnol. 1998, 49, 72–76. [Google Scholar] [CrossRef]
- Gupta, A.; Barrow, C.J.; Puri, M. Omega-3 biotechnology: Thraustochytrids as a novel source of omega-3 oils. Biotechnol. Adv. 2012, 30, 1733–1745. [Google Scholar] [CrossRef]
- Nakazawa, A.; Matsuura, H.; Kose, R.; Ito, K.; Ueda, M.; Honda, D.; Inouye, I.; Kaya, K.; Watanabe, M.M. Optimization of biomass and fatty acid production by Aurantiochytrium sp. Strain 4W-1b. Procedia Environ. Sci. 2012, 15, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zheng, Y.; Gao, H.; Xu, P.; Wang, M.; Li, A.; Miao, M.; Xie, X.; Deng, Y.; Zhou, H.; et al. Identification and characterization of Staphylococcus aureus strains with an incomplete hemolytic phenotype. Front. Cell. Infect. Microbiol. 2016, 6, 146. [Google Scholar] [CrossRef] [Green Version]
- Samie, A.; Ramalivhana, J.; Igumbor, E.; Obi, C. Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp. isolated from human diarrhoeal stools in Vhembe District, South Africa. J. Health Popul. Nutr. 2007, 25, 406–413. [Google Scholar]
- Satpute, S.K.; Bhawsar, B.D.; Dhakephalkar, P.K.; Chopade, B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria, Indian. J. Mar. Sci. 2008, 37, 243–250. [Google Scholar]
- Tayabali, A.F.; Nguyen, K.C.; Shwed, P.S.; Crosthwait, J.; Coleman, G.; Seligy, V.L. Comparison of the virulence potential of acinetobacter strains from clinical and environmental sources. PLoS ONE 2012, 7, e37024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrv, R.; Devaki, R.; Kandi, V. Comparison of hemagglutination and hemolytic activity of various bacterial clinical isolates against different human blood groups. Cureus 2016, 8, 489. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Vega, A.; Rosales-Mendoza, S.; Bañuelos-Hernandez, B.; Angulo, C. Prospects on the use of Schizochytrium sp. to develop oral vaccines. Front. Microbiol. 2018, 9, 2506. [Google Scholar] [CrossRef]
- Ravikumar, S.; Kathiresan, K. Marine Pharmacology; School of Marine Sciences, Alagappa University: Karaikudi, India, 2010; p. 215. ISBN 978-93-80400-12-9. [Google Scholar]
- Desbois, A.P.; Lawlor, A.C. Antibacterial activity of long-chain polyunsaturated fatty acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs 2013, 11, 4544–4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.Y.; Bajpai, V.K.; Kim, H.R.; Kang, S.C. Antibacterial activity of bioconverted eicosapentaenoic (EPA) and docosahexaenoic acid (DHA) against foodborne pathogenic bacteria. Int. J. Food Microbiol. 2007, 113, 233–236. [Google Scholar] [CrossRef]
- Narendran, R.; Kathiresan, K.; Rajendran, N.; Kandasamy, K. Antimicrobial activity of crude extracts from mangrove-derived Trichoderma species against human and fish pathogens. Biocatal. Agric. Biotechnol. 2016, 6, 189–194. [Google Scholar] [CrossRef]
- Yu, J.-H.; Wang, Y.; Sun, J.; Bian, F.; Chen, G.; Zhang, Y.; Bi, Y.-P.; Wu, Y.-J. Antioxidant activity of alcohol aqueous extracts of Crypthecodinium cohnii and Schizochytrium sp. J. Zhejiang Univ. Sci. B 2017, 18, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Tlili, N.; Khaldi, A.; Triki, S.; Munné-Bosch, S. Phenolic compounds and vitamin antioxidants of caper (Capparis spinosa). Plant Foods Hum. Nutr. 2010, 65, 260–265. [Google Scholar] [CrossRef]
- Jiang, J.; Shang, X.; Wang, H.; Xu, Y.-B.; Gao, Y.; Zhou, Q. Diagnostic value of contrast-enhanced ultrasound in thyroid nodules with calcification. Kaohsiung J. Med. Sci. 2015, 31, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Visudtiphole, V.; Phromson, M.; Tala, S.; Bunphimpapha, P.; Raweeratanapong, T.; Sittikankaew, K.; Arayamethakorn, S.; Preedanon, S.; Jangsutthivorawat, W.; Chaiyapechara, S.; et al. Aurantiochytrium limacinum BCC52274 improves growth, hypo-salinity tolerance and swimming strength of Penaeus vannamei post larvae. Aquaculture 2018, 495, 849–857. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Lee, M.-H.; Leong, Y.K.; Chang, J.-S.; Lee, D.-J. Biodiesel production from heterotrophic oleaginous microalga Thraustochytrium sp. BM2 with enhanced lipid accumulation using crude glycerol as alternative carbon source. Bioresour. Technol. 2020, 306, 123113. [Google Scholar] [CrossRef]
- Park, H.; Kwak, M.; Seo, J.; Ju, J.; Heo, S.; Park, S.; Hong, W. Enhanced production of carotenoids using a Thraustochytrid microalgal strain containing high levels of docosahexaenoic acid-rich oil. Bioprocess Biosyst. Eng. 2018, 41, 1355–1370. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Liefeldt, S.; Rova, U.; Christakopoulos, P.; Matsakas, L. Co-production of DHA and squalene by thraustochytrid from forest biomass. Sci. Rep. 2020, 10, 1992. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Xi, Y.; Mi, L.; Xiang, L.; Yanzhang, W.; Min, C.; Arthur, R.; Mark, T.; Junhuan, D.; Yi, Z. Investigation of composition, structure and bioactivity of extracellular polymeric substances from original and stress-induced strains of Thraustochytrium striatum, Carbohyd. Polymer 2018, 18, 01448617305216. [Google Scholar] [CrossRef]
- Mo, C.; Douek, J.; Rinkevich, B. Development of a PCR strategy for thraustochytrid identification based on 18S rDNA sequence. Mar. Biol. 2002, 140, 883–889. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A Simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Meyer-Rochow, V.B.; Nishimura, K.; Eguchi, E. Fatty acid composition and ultrastructure of photoreceptive membranes in the crayfish Procambarus clarkii under conditions of thermal and photic stress. J. Comp. Physiol. B 1997, 167, 1–8. [Google Scholar] [CrossRef]
- Buxton, R. Blood Agar Plates and Haemolysis Protocols. Am. Soc. Microbiol. 2005, 9. Available online: http://www.asmscience.org/content/education/protocol/protocol.2885 (accessed on 7 October 2021).
- Kalidasan, K.; Ravi, V.; Sunil, K.S.; Maheshwaran, M.L.; Kathiresan, K. Antimicrobial and anticoagulant activities of the spine of Stingray Himantura imbricata (Bloch & Schneider). J. Coast. Life Med. 2014, 2, 89–93. [Google Scholar] [CrossRef]
- Duan, X.J.; Zhang, W.W.; Li, X.M.; Wang, B.G. Evaluation of antioxidant property of extract and fractions obtained from a red alga, Polysiphonia urceolata. Food Chem. 2006, 95, 37–43. [Google Scholar] [CrossRef]








| Isolate No. | Mangrove Habitats | Colony Color | Colony Morphology | Ectoplasmic Network | Cell Wall | Amoeboid Cell | Binary Cell Division | Organism Name |
|---|---|---|---|---|---|---|---|---|
| ANVKK-01 | MB | Pale white | Large size colonies, binary cell partition, thin wall globes, or spherical shape of thallus. 2, 4, 8 cluster of vegetative cells, biflagellate zoospores | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-02 | CC | Pale white | Medium size colonies, oblong or sub-oblong shape cells, amoeboid cells present, internal proliferation with motile zoospore formation, presence of biflagellate zoospores | Absent | Thick cell wall | Amoeboid cell absent | Absent | Thraustochytrium sp. |
| ANVKK-03 | MJ, MB | Orange | Moderate size colonies, globes and sub globes thallus. Amoeboid cells present, binary cell division present | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium limacinum. |
| ANVKK-04 | GP | White | Medium size colonies, bi-partition cleavage, diads, tetrads, and different numbers vegetative cells present, present, globes, spherical, or pseudopodia thallus, reniform or ovoid shape zoospores | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-05 | BN | White | Large size colonies, spherical, or sub-spherical shape thallus, internal proliferation with motile zoospore formation, biflagellate zoospores present | Well developed | Thin cell wall | Amoeboid cell present | Absent | Thraustochytriidae sp. |
| ANVKK-06 | SG | Creamy orange | Small to large size colonies, globes and sub-globes thallus, binary cell division present, 2, 4, 7, and different numbers of vegetative cells present, Limaciform amoeboid cells present | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-07 | CT | Creamy white | Small size colonies, globes and sub-globes thallus, multiproliferous bodies, Zoosporangia contain large vesicles | Absent | Thick cell wall | Amoeboid cell present | Absent | Aurantiochytrium sp. |
| ANVKK-08 | WD | Creamy white | Moderate to very large size colonies, binary cell division present, 2, 4, and different numbers of vegetative cells present, Pseudopodia or spherical shape of cell, zoospores actively present | Un-developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-09 | CC | Pale white | Medium size colonies, globes or sub-globes, bi-partition cleavage, tetrads, ovoid shape zoospores | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-10 | MJ | Creamy white | Moderate size colonies, large vesicle, multiproliferous bodies, zoosporangia contain large vesicle, many active zoospores | Un-developed | Thick cell wall | Amoeboid cell present | Absent | Aurantiochytrium sp. |
| ANVKK-11 | CC | Pale white | Small to medium, globes, or sub-globes shape of cell, binary cell division present, diads, tetrads, and different numbers of cells present, many active amoeboid cells present | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| ANVKK-12 | CT | Creamy white | Medium size cell, spherical, or pseudopodia shape of cell, bi-partition cleavage, 2, 4, and different numbers of vegetative cells present, different shape of limaciform amoeboid cells present | Well developed | Thin cell wall | Amoeboid cell present | Present | Aurantiochytrium sp. |
| Isolates. | Growth Range of pH Tolerances | Optimum pH | Growth Range of Temperature Tolerances | Optimum Temperature | Growth range of Salinity Tolerances | Optimum Salinity | Blood Agar Hemolysis |
|---|---|---|---|---|---|---|---|
| ANVKK-01 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-02 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-03 | 5–9 | 6.5–7.5 | 15–35 | 25–30 | 5–90 | 25–30 | Negative |
| ANVKK-04 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-05 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-06 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-07 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-08 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 5–90 | 25–30 | Negative |
| ANVKK-09 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-10 | 5–9 | 6.5–7.5 | 10–35 | 25–30 | 5–90 | 25–30 | Negative |
| ANVKK-11 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| ANVKK-12 | 4–10 | 6.5–7.5 | 10–35 | 25–30 | 0–100 | 25–30 | Negative |
| Fatty Acids | ANVKK-01 | ANVKK-02 | ANVKK-03 | ANVKK-04 | ANVKK-05 | ANVKK-06 | ANVKK-07 | ANVKK-08 | ANVKK-09 | ANVKK-10 | ANVKK-11 | ANVKK-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C4:0 | 0.24 | 0 | 0 | 0 | 0 | 0 | 0 | 0.07 | 0.64 | 0.01 | 0.56 | 0 |
| C7:0 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 | 0.5 | 0 | 0 | 0.14 |
| C11:0 | 0.50 | 0 | 0 | 0.23 | 3.31 | 0.15 | 0 | 1.46 | 1.21 | 0.3 | 0.93 | 0.12 |
| C12:0 | 0.41 | 0 | 0 | 0.61 | 2.28 | 0.19 | 0.21 | 2.17 | 0.43 | 0.02 | 0.61 | 0.64 |
| C13:0 | 6.81 | 0 | 0 | 0.09 | 7.03 | 2.04 | 2.23 | 5.33 | 6.54 | 0.43 | 6.05 | 8.02 |
| C14:0 | 12.62 | 27.59 | 0 | 2.59 | 1.84 | 0.68 | 1.37 | 8.45 | 5.39 | 8.68 | 3.87 | 2.23 |
| C14:1 | 2.4 | 0 | 0 | 0.11 | 11.28 | 5.37 | 10.44 | 0.35 | 2.64 | 0.03 | 9.12 | 0.26 |
| C15:0 | 0 | 0 | 0 | 18.02 | 29.1 | 2.23 | 16.63 | 29.56 | 28.77 | 0.74 | 0 | 0 |
| C16:0 | 46.11 | 39.71 | 41.05 | 8.32 | 3.18 | 19.63 | 1.39 | 0.13 | 6.55 | 28.17 | 27.76 | 32.76 |
| C16:1 | 5.33 | 14.11 | 23.68 | 0.38 | 4.15 | 5.04 | 7.55 | 6.67 | 4.63 | 1.55 | 2.53 | 0.82 |
| C16:2 | 0.26 | 0 | 0 | 1.15 | 8.75 | 0.16 | 0.03 | 0.04 | 0 | 0.17 | 0 | 0.43 |
| C16:3 | 0.52 | 0 | 0 | 0.08 | 0 | 0.06 | 0 | 0 | 0.42 | 0.07 | 1.38 | 0 |
| C16:4 | 0 | 0 | 0 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 | 0 | 0 |
| C17:1 | 2.41 | 8.56 | 1.11 | 0.07 | 0 | 1.78 | 4.53 | 0.79 | 4.78 | 0.56 | 3.56 | 3.89 |
| C17:3 | 0.44 | 0 | 0.34 | 0.98 | 0 | 0.21 | 0.08 | 0.17 | 0.08 | 0.11 | 0.76 | 0.38 |
| C18:0 | 0.86 | 0 | 0 | 0 | 0 | 0 | 2.13 | 3.32 | 0 | 2.56 | 0 | 1.61 |
| C18:1 | 1.55 | 0.98 | 1.51 | 2.06 | 3.61 | 0.09 | 0.22 | 0.39 | 0.1 | 0.14 | 3.41 | 0.21 |
| C18:2 | 0 | 0 | 0 | 0.16 | 0.78 | 0.07 | 0 | 0 | 0 | 1.08 | 0 | 0.29 |
| C18:3 | 0 | 0 | 0 | 0 | 0 | 2.05 | 2.41 | 0 | 1.03 | 0 | 0.98 | 1.3 |
| C20:0 | 2.34 | 5.69 | 0 | 0.98 | 1.21 | 0.26 | 0.34 | 1.08 | 0.67 | 0.65 | 2.59 | 3.64 |
| C20:3 | 0.28 | 0.23 | 0 | 2.47 | 4.67 | 0.08 | 0.65 | 0 | 0 | 0.34 | 0 | 0.33 |
| C20:4 | 1.04 | 0 | 0.44 | 0.34 | 0 | 0.4 | 0.75 | 3.92 | 0.51 | 0.87 | 2.61 | 3.94 |
| C20:5 | 0.34 | 0 | 0.37 | 11.03 | 0 | 3.67 | 1.91 | 8.16 | 3.2 | 4.41 | 0.98 | 6.92 |
| C21:5 | 0.56 | 0 | 0.24 | 1.61 | 0 | 0.02 | 0.19 | 0 | 0 | 0.08 | 0 | 0 |
| C22:5 | 2.91 | 0.76 | 1.07 | 4.65 | 2.73 | 8.21 | 8.65 | 5.16 | 8.01 | 6.89 | 5.67 | 0 |
| C22:6 | 12.98 | 2.37 | 30.19 | 44.07 | 16.08 | 47.19 | 38.29 | 22.66 | 23.9 | 42.14 | 26.63 | 32.07 |
| Clinical Pathogens | Zone of Inhibition (mm) (Mean ± S.D) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human Bacterial Pathogens | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | A (C) |
| E. coli | 6.32 ± 0.5 | 11.03 ± 0.4 | 7.81 ± 1.0 | - | - | 5.96 ± 1.52 | 4.34 ± 0.5 | 12.55 ± 0.6 | - | 11.06 ± 0.7 | 9.10 ± 1.0 | 16.63 ± 1.5 | 22.11 ± 0.5 |
| S. aureus | 8.06 ± 0.3 | - | 18.69 ± 1.2 | 12.58 ± 0.4 | 9.11 ± 0.2 | 17.58 ± 0.6 | 11.32 ± 1.5 | - | 15 ± 1.5 | 6.19 ± 1.15 | 16.24 ± 1.5 | 10.03 ± 0.5 | 20.27 ± 0.8 |
| K. pneumonia | - | 12.10 ± 0.6 | - | 11.73 ± 1.5 | 14.33 ± 1.1 | - | - | 11.45 ± 1.0 | 16.34 ± 0.7 | 5.57 ± 1.5 | - | 4.56 ± 1.5 | 21.09 ± 0.4 |
| V. cholera | 4.32 ± 0.2 | - | 13.06 ± 0.5 | - | 7.89 ± 0.3 | 5.03 ± 1.5 | 14 ± 1.2 | - | 7.08 ± 0.2 | 10.56 ± 0.5 | 7.35 ± 0.8 | - | 19.21 ± 1.5 |
| B. subtilis | - | 8.00 ± 0.7 | 13.66 ± 1.5 | 10.97 ± 0.2 | - | 17.66 ± 0.5 | 15.23 ± 0.2 | 14.11 ± 2.0 | - | 11 ± 1.0 | 14.51 ± 1.5 | 11.40 ± 1.0 | 21.12 ± 0.5 |
| Fish Pathogens | |||||||||||||
| V. alginolyticus | 9.02 ± 0.8 | 4.15 ± 0.4 | 4.67 ± 1.52 | 9.24 ± 0.3 | - | 7.23 ± 0.51 | 11.87 ± 2.0 | - | 14.33 ± 0.5 | - | 6.79 ± 1.15 | - | 16.34 ± 2.0 |
| V. parahaemolyticus | - | 7.33 ± 1.1 | - | 17.00 ± 1.5 | 7.66 ± 0.5 | 7.89 ± 0.55 | - | 9.33 ± 0.6 | 7.63 ± 0.3 | 18.31 ± 1.0 | - | 10.34 ± 0.5 | 21.23 ± 0.5 |
| V. campilli | 14.58 ± 0.4 | - | 9.03 ± 0.57 | 10.87 ± 1.3 | 5.56 ± 1.12 | - | 15.33 ± 1.5 | 10.66 ± 0.5 | - | 7.78 ± 1.52 | 5.78 ± 2.0 | 7.33 ± 1.5 | 18 ± 1.52 |
| V. harveyi | - | 14.67 ± 0.5 | 8.97 ± 0.6 | - | - | 5.09 ± 1.5 | 9.87 ± 1.1 | - | 5.78 ± 1.5 | 16.66 ± 1.1 | 11 ± 0.5 | 4.66 ± 1.12 | 19 ± 0.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalidasan, K.; Vinithkumar, N.V.; Peter, D.M.; Dharani, G.; Dufossé, L. Thraustochytrids of Mangrove Habitats from Andaman Islands: Species Diversity, PUFA Profiles and Biotechnological Potential. Mar. Drugs 2021, 19, 571. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100571
Kalidasan K, Vinithkumar NV, Peter DM, Dharani G, Dufossé L. Thraustochytrids of Mangrove Habitats from Andaman Islands: Species Diversity, PUFA Profiles and Biotechnological Potential. Marine Drugs. 2021; 19(10):571. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100571
Chicago/Turabian StyleKalidasan, Kaliyamoorthy, Nambali Valsalan Vinithkumar, Dhassiah Magesh Peter, Gopal Dharani, and Laurent Dufossé. 2021. "Thraustochytrids of Mangrove Habitats from Andaman Islands: Species Diversity, PUFA Profiles and Biotechnological Potential" Marine Drugs 19, no. 10: 571. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100571