Oceanapiside, a Marine Natural Product, Targets the Sphingolipid Pathway of Fluconazole-Resistant Candida glabrata
Abstract
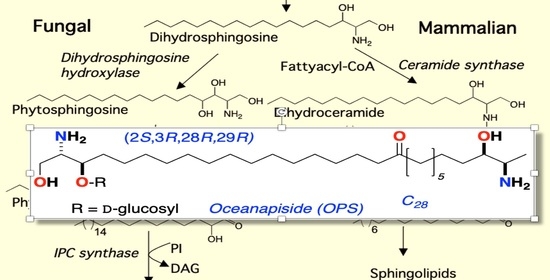
:1. Introduction
2. Results and Discussion
2.1. Effect of Oceanapiside (OPS) on Cell Viability in C. glabrata
2.2. Oceanapiside, OPS Affects Budding, Polarized Growth, and Actin Assembly in C. glabrata
2.3. Phytosphingosine Reverses the Antifungal Activity of Oceanapiside (OPS)
2.4. Oceanapiside (OPS) Induces the Accumulation of Phytosphingosine
3. Materials and Methods
3.1. Animal Materials and Natural Products
3.2. Media, Yeast Strain, and Culture Conditions
3.3. Time-Dependent Susceptibility Assay
3.4. Live and Dead Susceptibility Assay
3.5. Morphological Studies of Yeast Cells
3.6. Actin and Nucleus Staining of C. glabrata
3.7. Microscopy and Imaging Analysis
3.8. Long-chain Bases(LCBs) Rescue Experiment
3.9. Preparation of Cell Extracts
3.10. High Performance Liquid Chromatography and Electrospray Ionization Mass Spectrometric (HPLC-ESI-MS) Analysis of LCBs and Sphingolipids
3.11. NMR Spectrscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cacaci, M.; Menchinelli, G.; Torelli, R.; Sanglard, D.; Sanguinetti, M.; Posteraro, B. New Data on the In Vitro Activity of Fenticonazole against Fluconazole-Resistant Candida Species. Antimicrob. Agents Chemother. 2020, 64, 12. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, G.; Ebrahimi-Rad, M.; Mousavi, S.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Emergence of non-Candida albicans species: Epidemiology, phylogeny and fluconazole susceptibility profile. J. Med. Mycol. 2018, 28, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hokken, M.W.; Zwaan, B.; Melchers, W.; Verweij, P. Facilitators of adaptation and antifungal resistance mechanisms in clinically relevant fungi. Fungal Genet. Biol. 2019, 132, 103254. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.H.; Peacock, J.E.; Morris, A.J.; Tanner, D.C.; Snydman, D.R.; Wagener, M.M.; Rinaldi, M.G.; Yu, V.L. The changing face of candidemia: Emergence of non-Candida albicans species and antifungal resistance. Am. J. Med. 1996, 100, 617–623. [Google Scholar] [CrossRef]
- Vazquez, J.A.; Sobel, J.D.; Peng, G.; Steele-Moore, L.; Schuman, P.; Holloway, W.; Neaton, J.D. Evolution of vaginal Candida species recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis: The emergence of Candida glabrata? Terry Beirn Community Programs for Clinical Research in AIDS (CPCRA). Clin. Infect. Dis. 1999, 28, 1025–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gale, A.N.; Sakhawala, R.M.; Levitan, A.; Sharan, R.; Berman, J.; Timp, W.; Cunningham, K.W. Identification of Essential Genes and Fluconazole Susceptibility Genes in Candida glabrata by Profiling Hermes Transposon Insertions. G3: Genes Genomes Genet. 2020, 10, 3859–3870. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diekema, D.; Arbefeville, S.; Boyken, L.; Kroeger, J.; Pfaller, M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn. Microbiol. Infect. Dis. 2012, 73, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Fidel, P.L.; Vazquez, J.A.; Sobel, J.D. Candida glabrata: Review of Epidemiology, Pathogenesis, and Clinical Disease with Comparison toC. albicans. Clin. Microbiol. Rev. 1999, 12, 80–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchiesi, F.; Spreghini, E.; Maracci, M.; Fothergill, A.W.; Baldassarri, I.; Rinaldi, M.G.; Scalise, G. In Vitro Activities of Voriconazole in Combination with Three Other Antifungal Agents against Candida glabrata. Antimicrob. Agents Chemother. 2004, 48, 3317–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolotin-Fukuhara, M.; Fairhead, C. Editorial: Candida glabrata, the other yeast pathogen. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, C.F.; Silva, S.C.; Henriques, M. Candida glabrata: A review of its features and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 673–688. [Google Scholar] [CrossRef] [Green Version]
- Bossche, H.V.; Willemsens, G.; Marichal, P. Anti-Candida Drugs—The Biochemical Basis for Their Activity. CRC Crit. Rev. Microbiol. 1987, 15, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ji, X. Analysis of the microbial species, antimicrobial sensitivity and drug resistance in 2652 patients of nursing hospital. Heliyon 2020, 6, 03965. [Google Scholar] [CrossRef] [PubMed]
- Shadkchan, Y.; Segal, E. Antifungal activity of amphotericin B–lipid admixtures in experimental systemic candidosis in naive mice. J. Antimicrob. Chemother. 1999, 44, 787–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagiec, M.M.; Young, C.L.; Zaworski, P.G.; Kobayashi, S.D. Yeast sphingolipid bypass mutants as indicators of antifungal agents selectively targeting sphingolipid synthesis. Biochem. Biophys. Res. Commun. 2003, 307, 369–374. [Google Scholar] [CrossRef]
- Nagiec, M.M.; Nagiec, E.E.; Baltisberger, J.A.; Wells, G.B.; Lester, R.L.; Dickson, R.C. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 1997, 272, 9809–9817. [Google Scholar] [CrossRef] [Green Version]
- Healey, K.R.; Challa, K.K.; Edlind, T.D.; Katiyar, S.K. Sphingolipids Mediate Differential Echinocandin Susceptibility in Candida albicans and Aspergillus nidulans. Antimicrob. Agents Chemother. 2015, 59, 3377–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.; Singh, A.; Kumari, S.; Kumar, P.; Wasi, M.; Mondal, A.K.; Rudramurthy, S.M.; Chakrabarti, A.; Gaur, N.A.; Gow, N.A.; et al. Sphingolipidomics of drug resistant Candida auris clinical isolates reveal distinct sphingolipid species signatures. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158815. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Katiyar, S.K.; Raj, S.; Edlind, T.D. CRS-MIS in Candida glabrata: Sphingolipids modulate echinocandin-Fks interaction. Mol. Microbiol. 2012, 86, 303–313. [Google Scholar] [CrossRef] [Green Version]
- McEvoy, K.; Normile, T.G.; Del Poeta, M. Antifungal Drug Development: Targeting the Fungal Sphingolipid Pathway. J. Fungi 2020, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.M.; Del Poeta, M. Fungal sphingolipids: Role in the regulation of virulence and potential as targets for future antifungal therapies. Expert Rev. Anti-Infective Ther. 2020, 18, 1083–1092. [Google Scholar] [CrossRef]
- Mor, V.; Rella, A.; Farnoud, A.M.; Singh, A.; Munshi, M.; Bryan, A.M.; Naseem, S.; Konopka, J.B.; Ojima, I.; E Bullesbach, E.; et al. Identification of a New Class of Antifungals Targeting the Synthesis of Fungal Sphingolipids. mBio 2015, 6, e00647-15. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.C. Sphingolipid functions in Saccharomyces cerevisiae: Comparison to mammals. Annu. Rev. Biochem. 1998, 67, 27–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.C.; Lester, R.L. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1999, 1438, 305–321. [Google Scholar] [CrossRef]
- Obeid, L.M.; Okamoto, Y.; Mao, C. Yeast sphingolipids: Metabolism and biology. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2002, 1585, 163–171. [Google Scholar] [CrossRef]
- Guillas, I.; Kirchman, P.A.; Chuard, R.; Pfefferli, M.; Jiang, J.C.; Jazwinski, S.M.; Conzelmann, A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001, 20, 2655–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schorling, S.; Vallée, B.; Barz, W.P.; Riezman, H.; Oesterhelt, D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001, 12, 3417–3427. [Google Scholar] [CrossRef] [PubMed]
- Haak, D.; Gable, K.; Beeler, T.; Dunn, T. Hydroxylation of Saccharomyces cerevisiae Ceramides Requires Sur2p and Scs7p. J. Biol. Chem. 1997, 272, 29704–29710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montefusco, D.J.; Matmati, N.; Hannun, Y.A. The yeast sphingolipid signaling landscape. Chem. Phys. Lipids 2014, 177, 26–40. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, S.; Greenberg, M.L. Harnessing the power of yeast to elucidate the role of sphingolipids in metabolic and signaling processes pertinent to psychiatric disorders. Clin. Lipidol. 2014, 9, 533–551. [Google Scholar] [CrossRef] [Green Version]
- Nicholas, G.M.; Hong, T.W.; Molinski, T.F.; Lerch, M.L.; Cancilla, M.T.; Lebrilla, C.B. Oceanapiside, an antifungal bis- α,ω--amino alcohol glycoside from the marine sponge Oceanapia phillipensis. J. Nat. Prod. 1999, 62, 1678–1681. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, G.M.; Molinski, T.F. Enantiodivergent Biosynthesis of the Dimeric Sphingolipid Oceanapiside from the Marine Sponge Oceanapia phillipensis. Determination of Remote Stereochemistry. J. Am. Chem. Soc. 2010, 31, 4011–4019. [Google Scholar] [CrossRef]
- Nicholas, G.M.; Li, R.; Macmillan, J.B.; Molinski, T.F. Antifungal activity of bifunctional sphingolipids. intramolecular synergism within long-chain α,ω-bis-aminoalcohols. Bioorganic Med. Chem. Lett. 2002, 12, 2159–2162. [Google Scholar] [CrossRef]
- Millard, P.J.; Roth, B.L.; Thi, H.P.; Yue, S.T.; Haugland, R.P. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 1997, 63, 2897–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucsera, J.; Yarita, K.; Takeo, K. Simple detection method for distinguishing dead and living yeast colonies. J. Microbiol. Methods 2000, 41, 19–21. [Google Scholar] [CrossRef]
- Friant, S.; Lombardi, R.; Schmelzle, T.; Hall, M.N.; Riezman, H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001, 20, 6783–6792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Park, T.-S.; Fischl, A.S.; Ye, X.S. Cell Cycle Progression and Cell Polarity Require Sphingolipid Biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 2001, 21, 6198–6209. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Takesako, K.; Kato, I.; Yamaguchi, H. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob. Agents. Chemother. 1997, 41, 672–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.C.; Lester, R.L. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2002, 1583, 13–25. [Google Scholar] [CrossRef]
- Dickson, R.C.; Sumanasekera, C.; Lester, R.L. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 2006, 45, 447–465. [Google Scholar] [CrossRef]
- Dickson, R.C. Roles for Sphingolipids in Saccharomyces cerevisiae. Retin. Degener. Dis. 2010, 688, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Karpova, T.S.; McNally, J.G.; Moltz, S.L.; Cooper, J.A. Assembly and Function of the Actin Cytoskeleton of Yeast: Relationships between Cables and Patches. J. Cell Biol. 1998, 142, 1501–1517. [Google Scholar] [CrossRef]
- Gihana, G.M.; Cross-Najafi, A.A.; Lacefield, S. The mitotic exit network regulates the spatiotemporal activity of Cdc42 to maintain cell size. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef] [PubMed]
- Garabedian, M.V.; Wirshing, A.; Vakhrusheva, A.; Turegun, B.; Sokolova, O.S.; Goode, B.L. A septin-Hof1 scaffold at the yeast bud neck binds and organizes actin cables. Mol. Biol. Cell 2020, 31, 1988–2001. [Google Scholar] [CrossRef]
- Akram, Z.; Ahmed, I.; Mack, H.; Kaur, R.; Silva, R.C.; Castilho, B.A.; Friant, S.; Sattlegger, E.; Munn, A.L. Yeast as a Model to Understand Actin-Mediated Cellular Functions in Mammals—Illustrated with Four Actin Cytoskeleton Proteins. Cells 2020, 9, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buede, R.; Rinker-Schaffer, C.; Pinto, W.J.; Lester, R.L.; Dickson, R.C. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 1991, 173, 4325–4332. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Norred, W.P.; Bacon, C.W.; Riley, R.T.; Merrill, A.H. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 1991, 266, 14486–14490. [Google Scholar] [CrossRef]
- Molinski, T. Antifungal Compounds from Marine Organisms. Curr. Med. Chem. Anti-Infective Agents 2004, 3, 197–220. [Google Scholar] [CrossRef]
- Lester, R.L.; Dickson, R.C. High-Performance Liquid Chromatography Analysis of Molecular Species of Sphingolipid-Related Long Chain Bases and Long Chain Base Phosphates in Saccharomyces cerevisiae after Derivatization with 6-Aminoquinolyl-N-hydroxysuccinimidyl Carbamate. Anal. Biochem. 2001, 298, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; Krohn, R.; Hermanson, G.; Mallia, A.; Gartner, F.; Provenzano, M.; Fujimoto, E.; Goeke, N.; Olson, B.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalisay, D.S.; Rogers, E.W.; Molinski, T.F. Oceanapiside, a Marine Natural Product, Targets the Sphingolipid Pathway of Fluconazole-Resistant Candida glabrata. Mar. Drugs 2021, 19, 126. https://0-doi-org.brum.beds.ac.uk/10.3390/md19030126
Dalisay DS, Rogers EW, Molinski TF. Oceanapiside, a Marine Natural Product, Targets the Sphingolipid Pathway of Fluconazole-Resistant Candida glabrata. Marine Drugs. 2021; 19(3):126. https://0-doi-org.brum.beds.ac.uk/10.3390/md19030126
Chicago/Turabian StyleDalisay, Doralyn S., Evan W. Rogers, and Tadeusz F. Molinski. 2021. "Oceanapiside, a Marine Natural Product, Targets the Sphingolipid Pathway of Fluconazole-Resistant Candida glabrata" Marine Drugs 19, no. 3: 126. https://0-doi-org.brum.beds.ac.uk/10.3390/md19030126