Untargeted Metabolomics Approach for the Discovery of Environment-Related Pyran-2-Ones Chemodiversity in a Marine-Sourced Penicillium restrictum
Abstract
:1. Introduction
2. Results
2.1. Effects of Culture Medium Components on the Metabolome of P. restrictum
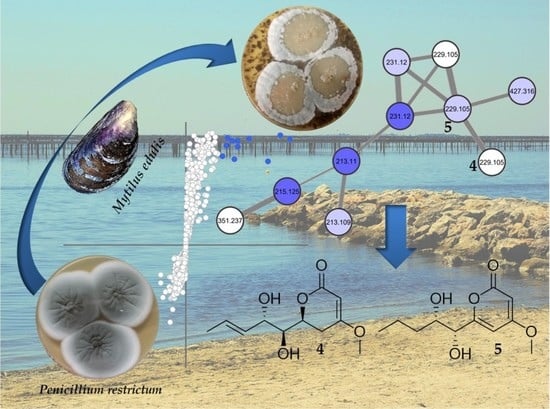
2.2. Molecular Networking of P. restrictum MES-SSW Extract
2.3. Isolation and Structure Elucidation of Pyran-2-ones
2.4. Biological Evaluation of Isolated Compounds
2.5. PTP1B Docking
3. Discussion
4. Materials and Methods
4.1. Fungal Material
4.2. Culture Media Preparation
4.3. Fermentation and Extraction for OSMAC Approach
4.4. HPLC-MS Analyses
4.5. Data Processing
4.6. Statistical Analysis
4.7. Molecular Networking
4.8. Annotation of MS Features
4.9. Isolation and Identification of Specialized Metabolites
4.10. Computational Details
4.11. Cytotoxicity Assays
4.12. Antimicrobial Assay
4.13. Antileishmanial Activity on Leishmania infantum Axenic Amastigotes
4.14. Docking
4.15. Electronic Circular Dichroism Measurements
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gosling, E. Phylogeny and evolution of bivalve molluscs. In Marine Bivalve Molluscs; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 1–11. [Google Scholar]
- Pereira, C.S.; Possas, C.D.A.; Viana, C.M.; Rodrigues, D.D.P. Características de Vibrio parahaemolyticus isolados de mexilhões (Perna perna) comercializados em Niterói, Rio de Janeiro. Rev. Soc. Bras. Med. Trop. 2007, 40, 56–59. [Google Scholar] [CrossRef] [Green Version]
- Leal, M.C.M.R.; Cammarota, M.C.; Freire, D.M.G.; Sant’Anna, G.L., Jr. Hydrolytic enzymes as coadjuvants in the anaerobic treatment of dairy wastewaters. Braz. J. Chem. Eng. 2002, 19, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Potasman, I.; Paz, A.; Odeh, M. Infectious outbreaks associated with bivalve shellfish consumption: A worldwide perspective. Clin. Infect. Dis. 2002, 35, 921–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lévesque, B.; Barthe, C.; Dixon, B.R.D.R.; Parrington, L.J.P.J.; Martin, D.; Doidge, B.; Proulx, J.-F.-F.; Murphy, D. Microbiological quality of blue mussels (Mytilus edulis) in Nunavik, Quebec: A pilot study. Can. J. Microbiol. 2010, 56, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Sallenave-Namont, C.; Pouchus, Y.F.; Robiou du Pont, T.; Lassus, P.; Verbist, J.F. Toxigenic saprophytic fungi in marine shellfish farming areas. Mycopathologia 2000, 149, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, L.V.; Vysotskaya, M.A. Filamentous fungi associated with bivalve mollusks from polluted biotopes of Ussuriiskii bay, sea of Japan. Russ. J. Mar. Biol. 2005, 31, 382–385. [Google Scholar] [CrossRef]
- Santos, A.; Hauser-Davis, R.A.; Santos, M.J.S.; De Simone, S.G. Potentially toxic filamentous fungi associated to the economically important Nodipecten nodosus (Linnaeus, 1758) scallop farmed in southeastern Rio de Janeiro, Brazil. Mar. Pollut. Bull. 2017, 115, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.B.G.; Pang, K.-L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Div. 2019, 96, 347–433. [Google Scholar] [CrossRef]
- Spiteller, P. Chemical ecology of fungi. Nat. Prod. Rep. 2015, 32, 971–993. [Google Scholar] [CrossRef]
- Rédou, V.; Vallet, M.; Meslet-Cladière, L.; Kumar, A.; Pang, K.-L.; Pouchus, Y.F.; Barbier, G.; Grovel, O.; Bertrand, S.; Prado, S.; et al. Chapter 4. Marine fungi. In The Marine Microbiome—An Untold Resource of Biodiversity and Biotechnological Potential, 1st ed.; Stal, L.J., Cretoiu, M.S., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 99–154. [Google Scholar]
- Antipova, T.V.; Zhelifonova, V.P.; Baskunov, B.P.; Kochkina, G.A.; Ozerskaya, S.M.; Kozlovskii, A.G. Exometabolites the Penicillium Fungi isolated from various high-latitude ecosystems. Microbiology 2018, 87, 642–651. [Google Scholar] [CrossRef]
- Vansteelandt, M.; Kerzaon, I.; Blanchet, E.; Fossi Tankoua, O.; Robiou Du Pont, T.; Joubert, Y.; Monteau, F.; Le Bizec, B.; Frisvad, J.C.; Pouchus, Y.F.; et al. Patulin and secondary metabolite production by marine-derived Penicillium strains. Fungal Biol. 2012, 116, 954–961. [Google Scholar] [CrossRef]
- Petit, K.E.; Mondeguer, F.; Roquebert, M.F.; Biard, J.F.; Pouchus, Y.F. Detection of griseofulvin in a marine strain of Penicillium waksmanii by ion trap mass spectrometry. J. Microbiol. Methods 2004, 58, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matallah-Boutiba, A.; Ruiz, N.; Sallenave-Namont, C.; Grovel, O.; Amiard, J.-C.; Pouchus, Y.F.; Boutiba, Z. Screening for toxigenic marine-derived fungi in Algerian mussels and their immediate environment. Aquaculture 2012, 342–343, 75–79. [Google Scholar] [CrossRef]
- Geiger, M.; Guitton, Y.; Vansteelandt, M.; Kerzaon, I.; Blanchet, E.; Robiou du Pont, T.; Frisvad, J.C.; Hess, P.; Pouchus, Y.F.; Grovel, O. Cytotoxicity and mycotoxin production of shellfish-derived Penicillium spp., a risk for shellfish consumers. Lett. Appl. Microbiol. 2013, 57, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overy, D.P.; Zidorn, C.; Petersen, B.O.; Duus, J.Ø.; Dalsgaard, P.W.; Larsen, T.O.; Phipps, R.K. Medium dependant production of corymbiferone a novel product from Penicillium hordei cultured on plant tissue agar. Tetrahedron Lett. 2005, 46, 3225–3228. [Google Scholar] [CrossRef]
- Overy, D.P.; Blunt, J.W. Corymbiferan lactones from Penicillium hordei: Stimulation of novel phenolic metabolites using plant tissue media. J. Nat. Prod. 2004, 67, 1850–1853. [Google Scholar] [CrossRef]
- Osman, M.; Stigloher, C.; Mueller, M.J.; Waller, F. An improved growth medium for enhanced inoculum production of the plant growth-promoting fungus Serendipita indica. Plant Methods 2020, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Cagigal, E.F.-M.; Sánchez, A.C. Influence of the culture media and the organic matter in the growth of Paxillus ammoniavirescens (Contu & Dessi). Mycobiology 2017, 45, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Overy, D.P.; Smedsgaard, J.; Frisvad, J.C.; Phipps, R.K.; Thrane, U. Host-derived media used as a predictor for low abundant, in planta metabolite production from necrotrophic fungi. J. Appl. Microbiol. 2006, 101, 1292–1300. [Google Scholar] [CrossRef]
- Medina, M.L.; Kiernan, U.A.; Francisco, W.A. Proteomic analysis of rutin-induced secreted proteins from Aspergillus flavus. Fungal Genet. Biol. 2004, 41, 327–335. [Google Scholar] [CrossRef]
- Stajic, M.; Persky, L.; Cohen, E.; Hadar, Y.; Brceski, I.; Wasser, S.P.; Nevo, E. Screening of laccase, manganese peroxidase, and versatile peroxidase activities of the genus Pleurotus in media with some raw plant materials as carbon sources. Appl. Biochem. Biotechnol. 2004, 117, 155–164. [Google Scholar] [CrossRef]
- Dhillon, B.; Feau, N.; Aerts, A.L.; Beauseigle, S.; Bernier, L.; Copeland, A.; Foster, A.; Gill, N.; Henrissat, B.; Herath, P.; et al. Horizontal gene transfer and gene dosage drives adaptation to wood colonization in a tree pathogen. Proc. Natl. Acad. Sci. USA 2015, 112, 3451–3456. [Google Scholar] [CrossRef] [Green Version]
- Torrado, A.; Vázquez, J.-A.; Prieto, M.-Á.; Fuciños, P.; Montemayor, M.-I.; Pastrana, L.; González, M.-P.; Murado, M.-Á. Amylase production by Aspergillus oryzae in a solid-state bioreactor with fed-batch operation using mussel processing wastewaters as feeding medium. J. Chem Technol. Biotechnol. 2013, 88, 226–236. [Google Scholar] [CrossRef]
- Sankaran, S.; Khanal, S.K.; Jasti, N.; Jin, B.; Pometto, A.L.; Van Leeuwen, J.H. Use of filamentous fungi for wastewater treatment and production of high Value fungal byproducts: A review. Crit. Rev. Environ. Sci. Technol. 2010, 40, 400–449. [Google Scholar] [CrossRef]
- Amado, I.R.; Vázquez, J.A. Mussel processing wastewater: A low-cost substrate for the production of astaxanthin by Xanthophyllomyces dendrorhous. Microb. Cell Factories 2015, 14, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastrana, L.M.; Gonzalez, M.P.; Murado, M.A. Production of gibberellic acid from mussel processing wastes in submerged batch culture. Bioresour. Technol. 1993, 45, 213–221. [Google Scholar] [CrossRef]
- González, M.P.; Siso, M.I.G.; Murado, M.A.; Pastrana, L.; Montemayor, M.I.; Mirón, J. Depuration and valuation of mussel-processing wastes. Characterization of amylolytic postincubates from different species grown on an effluent. Bioresour. Technol. 1992, 42, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Murado, M.A.; González, M.P.; Pastrana, L. Mussel processing wastes as a fermentation substrate. In Fisheries Processing; Springer: Boston, MA, USA, 1994; pp. 311–343. [Google Scholar]
- Nicoletti, R.; Stefano, M.D. Penicillium restrictum as an antagonist of plant pathogenic fungi. Dyn. Biochem. Process Biotechnol. Mol. Biol. 2012, 6, 61–69. [Google Scholar]
- Sankhala, R. Metabolic products of Penicillium restrictum. Indian J. Exp. Biol. 1968, 6, 57–58. [Google Scholar]
- Schwartz, R.E.; Dufresne, C.; Flor, J.E.; Kempf, A.J.; Wilson, K.E.; Lam, T.; Onishi, J.; Milligan, J.; Fromtling, R.A.; Abruzzo, G.K. Restricticin, a novel glycine-containing antifungal agent. J. Antibiot. 1991, 44, 463–471. [Google Scholar] [CrossRef]
- Hensens, O.D.; Wichmann, C.F.; Liesch, J.M.; Vanmiddlesworth, F.L.; Wilson, K.E.; Schwartz, R.E. Structure elucidation of restricticin, a novel antifungal agent from Penicillium restrictum. Tetrahedron 1991, 47, 3915–3924. [Google Scholar] [CrossRef]
- Raistrick, H.; Rice, F.A.H. 2,3-Dihydro-3,6-dihydroxy-2-methyl-4-pyrone and curvularin from Penicillium gilmanii. J. Chem. Soc. C Org. 1971, 3069. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Karwowski, J.P.; Humphrey, P.E.; Kohl, W.L.; Barlow, G.J.; Tanaka, S.K. Calbistrins, novel antifungal agents produced by Penicillium restrictum. I. Production, taxonomy of the producing organism and biological activity. J. Antibiot. 1993, 46, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Brill, G.M.; Chen, R.H.; Rasmussen, R.R.; Whittern, D.N.; McAlpine, J.B. Calbistrins, novel antifungal agents produced by Penicillium restrictum. II. Isolation and elucidation of structure. J. Antibiot. 1993, 46, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, A.; Jurado, M.; Rodríguez, M.; Núñez, F.; Córdoba, J.J. Characterization of molds from dry-cured meat products and their metabolites by micellar electrokinetic capillary electrophoresis and random amplified polymorphic DNA PCR. J. Food Prot. 2004, 67, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.P.T.; Roullier, C.; Boumard, M.-C.; Robiou du Pont, T.; Nazih, H.; Gallard, J.-F.; Pouchus, Y.F.; Beniddir, M.A.; Grovel, O. Metabolomics-driven discovery of meroterpenoids from a mussel-derived Penicillium ubiquetum. J. Nat. Prod. 2018, 81, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.I. Chapter 4—O- and S-heteroaromatic compounds. In Interpretation of the Ultraviolet Spectra of natural Products; Scott, A.I., Ed.; International Series of Monographs on Organic Chemistry; Pergamon: Oxford, UK, 1964; pp. 135–164. [Google Scholar]
- Collett, L.A.; Davies-Coleman, M.T.; Rivett, D.E.A. Naturally occurring 6-substituted 5,6-dihydro-α-pyrones. In Fortschritte der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products; Springer: Vienna, Austria, 1998; pp. 181–209. [Google Scholar]
- McGahren, W.J.; Ellestad, G.A.; Morton, G.O.; Kunstmann, M.P.; Mullen, P. New fungal lactone, LL-P880β, and a new pyrone, LL-880γ, from a Penicillium species. J. Org. Chem. 1973, 38, 3542–3544. [Google Scholar] [CrossRef]
- Gorst-Allman, C.; Steyn, P. Biosynthesis of 5, 6-dihydro-4-methoxy-2H-pyran-2-one in Penicillium italicum. S. Afr. J. Chem. 1983, 36, 83–84. [Google Scholar]
- Kimura, Y.; Hamasaki, T.; Nakajima, H. Stereochemistry and biological activities of LL-P880γ, a pestalotin analogue, produced by Penicillium citreo-viride. Agric. Biol. Chem. 1986, 50, 1649–1650. [Google Scholar] [CrossRef]
- Sakai, K.; Chiba, H.; Kaneto, R.; Sakamoto, M.; Okamura, K.; Tone, H. Mer-NF8054A and X, novel antifungal steroids, isolated from Aspergillus sp. J. Antibiot. 1994, 47, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Hao, X.; Li, S.; Jia, J.; Guan, Y.; Peng, Z.; Bi, H.; Xiao, C.; Cen, S.; et al. Broad-spectrum antiviral natural products from the marine-Derived Penicillium sp. IMB17-046. Molecules 2019, 24, 2821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, H.; Hara, M.; Yamashita, Y.; Ando, K.; Shuto, K. Paxisterol, a new analgesic sterol without anti-inflammation activity from Penicillium. J. Antibiot. 1988, 41, 409–410. [Google Scholar] [CrossRef] [Green Version]
- Okabe, M.; Sugita, T.; Kinoshita, K.; Koyama, K. Macrolides from a marine-derived fungus, Penicillium meleagrinum var. viridiflavum, showing synergistic effects with fluconazole against azole-resistant Candida albicans. J. Nat. Prod. 2016, 79, 1208–1212. [Google Scholar] [CrossRef]
- Gao, N.; Shang, Z.-C.; Yu, P.; Luo, J.; Jian, K.-L.; Kong, L.-Y.; Yang, M.-H. Alkaloids from the endophytic fungus Penicillium brefeldianum and their cytotoxic activities. Chin. Chem. Lett. 2017, 28, 1194–1199. [Google Scholar] [CrossRef]
- Ishimaru, T.; Harada, S.; Tsuboya, S. TAN-1446 Exhibiting Anti-Angiotensin II Activity and its Manufacture with Penicillium restrictum. Japan Patent JP 05041994, 23 February 1993. [Google Scholar]
- Larsen, T.O.; Frisvad, J.C.; Christophersen, C. Arabenoic acid (verrucolone), a major chemical indicator of Penicillium verrucosum. Biochem. Syst. Ecol. 1998, 26, 463–465. [Google Scholar] [CrossRef]
- Isaac, B.G.; Ayer, S.W.; Stonard, R.J. Arabenoic acid, a natural product herbicide of fungal origin. J. Antibiot. 1991, 44, 793–794. [Google Scholar] [CrossRef]
- Matsumura, M.; Miki, S.; Oyama, K.; Tsuchida, M.; Yamamoto, K. New Agricultural/Horticultural Bactericide. Japan Patent JP2007280064A, 29 October 2009. [Google Scholar]
- Nose, H.; Seki, A.; Yaguchi, T.; Hosoya, A.; Sasaki, T.; Hoshiko, S.; Shomura, T. PF1163A and B, new antifungal antibiotics produced by Penicillium sp. I. Taxonomy of producing strain, fermentation, isolation and biological activities. J. Antibiot. 2000, 53, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Nose, H.; Hosoya, A.; Yoshida, S.; Kawaguchi, M.; Watanabe, T.; Usui, T.; Ohtsuka, Y.; Shomura, T.; Takano, S.; et al. PF1163A and B, new antifungal antibiotics Produced by Penicilium sp. II. Physico-chemical properties and structure elucidation. J. Antibiot. 2000, 53, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellestad, G.A.; McGahren, W.J.; Kunstmann, M.P. Structure of a new fungal lactone, LL-P880α, from an unidentified Penicillium species. J. Org. Chem. 1972, 37, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Tamura, S. Isolation and structure of pestalotin, a gibberellin synergist from Pestalotia cryptomeriaecola. Agric. Biol. Chem. 1972, 36, 1925–1930. [Google Scholar] [CrossRef]
- Kimura, Y.; McGahren, W.J.; Suzuki, A.; Tamura, S. Structure of a new fungal pyrone, from an unidentified Penicillium sp. Agric. Biol. Chem. 1978, 42, 1625–1626. [Google Scholar] [CrossRef]
- Evidente, A.; Zonno, M.C.; Andolfi, A.; Troise, C.; Cimmino, A.; Vurro, M. Phytotoxic α-pyrones produced by Pestalotiopsis guepinii, the causal agent of hazelnut twig blight. J. Antibiot. 2012, 65, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Kim, S.; Liu, C.; Shim, S. Secondary metabolites produced by an endophytic fungus Pestalotiopsis sydowiana and their 20S proteasome inhibitory activities. Molecules 2016, 21, 944. [Google Scholar] [CrossRef] [Green Version]
- Pospisil, J.; Marko, I.E. Metathesis-based synthesis of 3-methoxy α,β-unsaturated lactones: Total synthesis of (R)-kavain and of the C1-C6 fragment of jerangolid D. Tetrahedron Lett. 2008, 49, 1523–1526. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, C.-X.; Yu, Y.; Wang, G.-Q.; Zheng, Q.-C.; Chen, G.-D.; Lian, Y.-Y.; Lin, F.; Guo, L.-D.; Gao, H. Nodulisporipyrones A–D, new bioactive α-pyrone derivatives from Nodulisporium sp. J. Asian Nat. Prod. Res. 2015, 17, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Kirihata, M.; Ohta, K.; Ichimoto, I.; Ueda, H. Total synthesis of (6S,1S,2R)-6-(1,2-Dihydroxypentyl)-4-methoxy-5,6-dihydropyrane-2-one (LL-P880β) and its C6-epimer, a fungal metabolite from Penicillium sp. Agric. Biol. Chem. 1990, 54, 2401–2405. [Google Scholar] [CrossRef]
- Rönsberg, D.; Debbab, A.; Mándi, A.; Wray, V.; Dai, H.; Kurtán, T.; Proksch, P.; Aly, A.H. Secondary metabolites from the endophytic fungus Pestalotiopsis virgatula isolated from the mangrove plant Sonneratia caseolaris. Tetrahedron Lett. 2013, 54, 3256–3259. [Google Scholar] [CrossRef] [Green Version]
- Kirihata, M.; Ohe, M.; Ichimoto, I.; Ueda, H. Stereoselective synthesis of unnatural stereoisomers of LL-P880β and LL-P880γ, pestalotin analogues from Penicillium sp. Biosci. Biotechnol. Biochem. 1992, 56, 1825–1828. [Google Scholar] [CrossRef]
- Kirihata, M.; Kamihisa, Y.; Ichimoto, I.; Ueda, H. Stereoselective synthesis of of (6R,1′R,2′S)- and (6S,1′R,2′S)-LL-P880β, stereoisomers of the fungal metabolite from Penicillium strains. Chem. Express 1992, 7, 837. [Google Scholar]
- Kirihata, M.; Ohe, M.; Ichimoto, I.; Kimura, Y. Synthesis and biological activity of LL-P880γ and its analogues. Biosci. Biotechnol. Biochem. 1996, 60, 677–679. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xu, C.-C.; Tang, H.; Su, L.; Chou, Y.; Soong, K.; Li, J.; Zhuang, C.-L.; Luo, Y.-P.; Zhang, W. Osteoclastogenesis inhibitory polyketides from the sponge-associated fungus Xylaria feejeensis. Chem. Biodivers. 2018, 15, e1800358. [Google Scholar] [CrossRef]
- Smith, S.G.; Goodman, J.M. Assigning stereochemistry to single diastereoisomers by GIAO NMR Calculation: The DP4 probability. J. Am. Chem. Soc. 2010, 132, 12946–12959. [Google Scholar] [CrossRef] [PubMed]
- Ermanis, K.; Parkes, K.E.B.; Agback, T.; Goodman, J.M. The optimal DFT approach in DP4 NMR structure analysis—pushing the limits of relative configuration elucidation. Org. Biomol. Chem. 2019, 17, 5886–5890. [Google Scholar] [CrossRef]
- Rao, K.V.; Reddy, G.C.S. A new reaction of patulin. J. Nat. Prod. 1989, 52, 1376–1378. [Google Scholar] [CrossRef]
- Ji, S.; Li, Z.; Song, W.; Wang, Y.; Liang, W.; Li, K.; Tang, S.; Wang, Q.; Qiao, X.; Zhou, D.; et al. Bioactive constituents of Glycyrrhiza uralensis (Licorice): Discovery of the effective components of a traditional herbal medicine. J. Nat. Prod. 2016, 79, 281–292. [Google Scholar] [CrossRef]
- Han, W.; Cai, J.; Zhong, W.; Xu, G.; Wang, F.; Tian, X.; Zhou, X.; Liu, Q.; Liu, Y.; Wang, J. Protein tyrosine phosphatase 1B (PTP1B) inhibitors from the deep-sea fungus Penicillium chrysogenum SCSIO 07007. Bioorganic Chem. 2020, 96, 103646. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Tonks, N.K. PTP1B: From the sidelines to the front lines! FEBS Lett. 2003, 546, 140–148. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Xie, L.; Yang, F.; Wang, W.; Zhou, Q.; Xiang, M.; Zhou, S.; Lv, W.; Jia, Y.; Pokhrel, L.; et al. Recent advance on PTP1B inhibitors and their biomedical applications. Eur. J. Med. Chem. 2020, 199, 112376. [Google Scholar] [CrossRef] [PubMed]
- Brandão, T.A.S.; Hengge, A.C.; Johnson, S.J. Insights into the reaction of protein-tyrosine phosphatase 1B. J. Biol. Chem. 2010, 285, 15874–15883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igboeli, H.A.; Marchbank, D.H.; Correa, H.; Overy, D.; Kerr, R.G. Discovery of primarolides A and B from marine fungus Asteromyces cruciatus using osmotic stress and treatment with suberoylanilide hydroxamic acid. Mar. Drugs 2019, 17, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auckloo, B.N.; Pan, C.; Akhter, N.; Wu, B.; Wu, X.; He, S. Stress-driven discovery of novel cryptic antibiotics from a marine fungus Penicillium sp. BB1122. Front. Microbiol. 2017, 8, 1450. [Google Scholar] [CrossRef] [Green Version]
- Overy, D.; Correa, H.; Roullier, C.; Chi, W.-C.; Pang, K.-L.; Rateb, M.; Ebel, R.; Shang, Z.; Capon, R.; Bills, G.; et al. Does osmotic stress affect natural product expression in fungi? Mar. Drugs 2017, 15, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lu, Z.; Sun, K.; Zhu, W. Effects of high salt stress on secondary metabolite production in the marine-derived fungus Spicaria elegans. Mar. Drugs 2011, 9, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Trisuwan, K.; Rukachaisirikul, V.; Borwornwiriyapan, K.; Phongpaichit, S.; Sakayaroj, J. Pyrone derivatives from the soil fungus Fusarium solani PSU-RSPG37. Phytochem. Lett. 2013, 6, 495–497. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.-M.; Meng, L.; Li, C.-S.; Gao, S.-S.; Shang, Z.; Proksch, P.; Huang, C.-G.; Wang, B.-G. Nigerapyrones A–H, α-pyrone derivatives from the marine mangrove-derived endophytic fungus Aspergillus niger MA-132. J. Nat. Prod. 2011, 74, 1787–1791. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Xu, L.-L.; Zhang, Y.; Lin, Z.-H.; Xia, T.; Yang, D.-F.; Chen, Y.-M.; Yang, X.-L. Three new α-pyrone derivatives from the plant endophytic fungus Penicillium ochrochloronthe and their antibacterial, antifungal, and cytotoxic activities. J. Asian Nat. Prod. Res. 2019, 21, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Evidente, A.; Cabras, A.; Maddau, L.; Serra, S.; Andolfi, A.; Motta, A. Viridepyronone, a new antifungal 6-substituted 2H-pyran-2-one produced byTrichoderma virid. J. Agric. Food. Chem. 2003, 51, 6957–6960. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.-S.; Ok, K.; Kim, Y.; Park, H.-D.; Byun, Y. Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 2913–2917. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Brameyer, S.; Kresovic, D.; Hitkova, I.; Kopp, Y.; Manske, C.; Schubert, K.; Bode, H.B.; Heermann, R. Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 2013, 9, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Servin, J.A.; Park, G.; Borkovich, K.A. Global analysis of serine/threonine and tyrosine protein phosphatase catalytic subunit genes in Neurospora crassa reveals interplay between phosphatases and the p38 mitogen-activated protein kinase. G3 Genes Genomes Genet. 2014, 4, 349–365. [Google Scholar] [CrossRef] [Green Version]
- Phaedra, E.; Athina, G.; Anthi, P. PTP1b Inhibition, A Promising Approach for the Treatment of Diabetes Type II. Curr. Top. Med. Chem. 2019, 19, 246–263. [Google Scholar] [CrossRef]
- Lee, D.-S.; Jang, J.-H.; Ko, W.; Kim, K.-S.; Sohn, J.; Kang, M.-S.; Ahn, J.; Kim, Y.-C.; Oh, H. PTP1B inhibitory and anti-Inflammatory effects of secondary metabolites isolated from the marine-derived fungus Penicillium sp. JF-55. Mar. Drugs 2013, 11, 1409–1426. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, C.; Schneider, K.; Bruntner, C.; Brown, R.; Jones, A.L.; Goodfellow, M.; Krämer, M.; Imhoff, J.F.; Nicholson, G.; Fiedler, H.-P.; et al. Albidopyrone, a new α-pyrone-containing metabolite from marine-derived Streptomyces sp. NTK 227. J. Antibiot. 2009, 62, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Helaly, S.; Schneider, K.; Nachtigall, J.; Vikineswary, S.; Tan, G.Y.A.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.-P. Gombapyrones, new α-pyrone metabolites produced by Streptomyces griseoruber Acta 3662. J. Antibiot. 2009, 62, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murgarella, M.; Puiu, D.; Novoa, B.; Figueras, A.; Posada, D.; Canchaya, C. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS ONE 2016, 11, e0151561. [Google Scholar] [CrossRef] [Green Version]
- Roullier, C.; Bertrand, S.; Blanchet, E.; Peigné, M.; Robiou du Pont, T.; Guitton, Y.; Pouchus, Y.F.; Grovel, O. Time dependency of biosynthetic pathways and chemodiversity: An LC-MS metabolomic study of marine-sourced Penicillium. Mar. Drugs 2016, 14, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.l.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bertrand, S.; Guitton, Y.; Roullier, C. Successes and pitfalls in automated dereplication strategy using liquid chromatography coupled to mass spectrometry data: A CASMI 2016 experience. Phytochem. Lett. 2017, 21, 297–305. [Google Scholar] [CrossRef]
- Böcker, S.; Letzel, M.C.; Lipták, Z.; Pervukhin, A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics 2009, 25, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A comprehensive species-metabolite relationship database. In Plant Metabolomics; Saito, K., Dixon, R., Willmitzer, L., Eds.; Biotechnology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 2006; Volume 57, pp. 165–181. Available online: http://kanaya.naist.jp/KNApSAcK/ (accessed on 28 June 2021).
- Wikler, M.A. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Ninth edition; (M2-A9); CLSI: Wayne, PA, USA, 2006. [Google Scholar]
- Clark, M.; Cramer, R.D.; Van Opdenbosch, N. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Erlanson, D.A.; McDowell, R.S.; He, M.M.; Randal, M.; Simmons, R.L.; Kung, J.; Waight, A.; Hansen, S.K. Discovery of a new phosphotyrosine mimetic for PTP1B using breakaway tethering. J. Am. Chem. Soc. 2003, 125, 5602–5603. [Google Scholar] [CrossRef]










| VIP | m/z | Rt (min) |
Observed Features (Ionic Species) |
Molecular Formula for M | ∆ppm |
UV-vis Absorption (λmax nm) | Putative Annotation | |
|---|---|---|---|---|---|---|---|---|
| SSW | 4.05 | 231.1225 | 6.52 | [M+H]+ | C11H18O5 | −3.24 | 238 | 2’-hydroxy pestalotin isomer |
| 253.1050 | [M+Na]+ | |||||||
| 3.46 | 411.3253 | 23.41 | [M+H]+ | C28H42O2 | −2.44 | n.d. | 3β-hydroxyergosta-8,14,24(28)-trien-7-one | |
| 3.43 | 472.3422 | 22.27 | [M+H]+ | C29H45NO4 | −1.02 | n.d. | N-demethylmelearoride A | |
| 494.3197 | [M+Na]+ | |||||||
| 3.15 | 486.3583 | 23.86 | [M+H]+ | C30H47NO4 | −0.07 | n.d. | Melearoride A | |
| 508.3407 | [M+Na]+ | |||||||
| 2.8 | 443.3150 | 23.87 | [M+H]+ | C28H42O4 | −2.56 | n.d. | Paxisterol | |
| 2.5 | 215.1281 | 9.08 | [M+H]+ | C11H18O4 | −1.09 | 237 | Pestalotin isomer | |
| 2.42 | 412.3335 | 23.28 | [M+H]+ | C26H41N3O | −1.53 | n.d. | - | |
| 2.34 | 427.3221 | 23.76 | [M+H]+ | C28H42O3 | 2.06 | n.d. | - | |
| 2.12 | 467.3157 | 19.62 | [M+Na]+ | C28H44O4 | 4.22 | n.d. | Antibiotic Mer-NF 8054A | |
| DW | 6.33 | 213.1139 | 9.82 | [M+H]+ | C11H16O4 | −2.78 | 279 | Dehydropestalotin isomer |
| 235.0972 | [M+Na]+ | |||||||
| 4.08 | 213.1132 | 10.03 | [M+H]+ | C11H16O4 | 2.42 | 237 | Pyran-2-one derivative | |
| 3.89 | 307.1577 | 14.92 | [M+H]+ | C17H22O5 | 10.26 | n.d. | Antibiotic TAN 1446A | |
| 3.88 | 213.1132 | 9.09 | [M+H]+ | C11H16O4 | −3.18 | 288 | Dehydropestalotin isomer | |
| 3.82 | 251.0899 | 11.56 | [M+Na]+ | C11H16O5 | 1.42 | 275 | LL-P880γ isomer | |
| 267.0638 | [M+K]+ | |||||||
| 3.72 | 129.0549 | 2.18 | [M+H]+ | C6H8O3 | −2.09 | 237 | 5,6-dihydro-4-methoxy-2H-pyran-2-one | |
| 3.24 | 431.2787 | 22.98 | [M+Na]+ | C24H40O5 | 3.14 | n.d. | Aspergillus acid A | |
| 2.97 | 515.2648 | 24.36 | [M+H]+ | C29H38O8 | 0.60 | n.d. | Citreohybridone B | |
| 2.9 | 147.0654 | 2.13 | [M+H]+ | C6H10O4 | −2.27 | 237 | 5-hydroxy-3-methoxy-2-pentenoic acid | |
| 2.79 | 379.3358 | 20.40 | [M+H]+ | C28H42 | −1.78 | n.d. | - | |
| 2.58 | 185.0845 | 5.88 | [M+H]+ | C12H11NO | 2.36 | n.d. | - | |
| 2.22 | 544.3654 | 18.59 | [M+H]+ | C32H49NO6 | 2.91 | n.d. | Pestalotiopin B | |
| 2.11 | 520.3413 | 16.45 | [M+ Na]+ | C31H47NO4 | 1.96 | n.d. | - |
| VIP | m/z | Rt (min) | Observed Features (Ionic Species) | Molecular Formula for M | ∆ppm | UV-vis Absorption (λmax nm) | Putative Annotation |
|---|---|---|---|---|---|---|---|
| 8.95 | 231.1225 | 6.52 | [M+H]+ | C11H18O5 | −3.24 | 238 | 2’-hydroxypestalotin isomer |
| 253.1050 | [M+Na]+ | ||||||
| 269.0800 | [M+K]+ | ||||||
| 483.2219 | [2M+Na]+ | ||||||
| 7.95 | 211.0581 | 6.58 | [M+Na]+ | C8H12O5 | −0.68 | 237 | Unknown pyran-2-one derivative |
| 5.45 | 213.1139 | 9.82 | [M+H]+ | C11H16O4 | 5.71 | 279 | Dehydroxypestalotin isomer |
| 4.59 | 129.0549 | 2.18 | [M+H]+ | C6H10O4 | −2.09 | 237 | 5,6-dihydro-4-methoxy-2H-pyran-2-one |
| 4.53 | 215.1281 | 9.08 | [M+H]+ | C11H18O4 | −1.09 | 237 | Pestalotin isomer |
| 237.1117 | [M+Na]+ | ||||||
| 3.57 | 251.0905 | 6.68 | [M+Na]+ | C11H16O5 | 5.8 | 275 | LL-P880γ isomer |
| 3.50 | 147.0654 | 2.13 | [M+H]+ | C6H10O4 | −4.99 | 237 | 5-hydroxy-3-methoxy-2-pentenoic acid |
| 3.24 | 251.0905 | 11.56 | [M+Na]+ | C11H16O5 | 1.82 | 275 | LL-P880γ isomer |
| Position | 1 | (6S,1’S,2’R)-LL-P880β | 2 | 3 | ||||
|---|---|---|---|---|---|---|---|---|
| δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | |
| 2 | 166.73 C | - | 166.58 C | 167.03 C | - | 167.18 C | - | |
| 3 | 89.95 CH | 5.16, s | 89.73 CH | 5.14 (d, 1.5) | 89.51 CH | 5.12, s | 89.55 CH | 5.11, s |
| 4 | 172.94 C | - | 173.40 C | 173.94 C | - | 173.68 C | - | |
| 5 | 28.84 CH2 | a. 2.8 (dd, 12.8, 16.7) b. 2.28 (dd, 3.7, 17.0) | 29.35 CH2 | a. 2.89 (ddd, 1.5, 12.9, 17.2) b. 2.32 (dd, 3.7, 16.9) | 29.49 CH2 | a. 3.05 (dd, 13.8, 16.7) b. 2.19 (dd, 2.9, 17.0) | 29.30 CH2 | a. 2.88 (dd, 13.9, 16.2) b. 2.31 (dd, 3.5, 17.2) |
| 6 | 76.21 CH | 4.51, m | 77.95 CH | 4.52 (dt, 4.0, 4.0, 12.9) | 75.41 CH | 4.76 (dt, 12.8, 2.3) | 77.88 CH | 4.5 (dt, 3.9, 4.2, 12.8) |
| 4-OCH3 | 56.18 | 3.77, s | 56.16 | 3.76, s | 56.16 | 3.76, s | 56.18 | 3.74, s |
| 1’ | 63.74 CH2 | a. 3.73 (dd, 4.6, 12.2) | 73.80 CH | 3.49 (dd, 2.9, 4.2) | 71.27 CH | 3.37 (dd, 2.0, 6.7) | 70.77 CH | 3.47 (t, 3.5, 3.9) |
| b. 3.91, m | ||||||||
| 2’ | - | - | 70.93 CH | 3.80, m | 74.13 CH | 3.80 (t, 7.1, 7.7) | 73.78 CH | 3.77 (t, 3.5, 4.3) |
| 3’ | - | - | 35.94 CH2 | 1.5–1.63, m | 35.71 CH2 | a. 1.66, m b. 1.5, m | 35.84 CH2 | 1.5–1.58, m |
| 4’ | - | - | 18.78 CH2 | 1.38–1.5, m | 18.78 CH2 | a. 1.58, m b. 1.4, m | 18.72 CH2 | 1.38–1.5, m |
| 5’ | - | - | 13.94 CH3 | 0.94 (t, 7.1) | 13.98 CH3 | 0.95 (t, 7.1) | 13.92 CH3 | 0.92 (t, 6.7) |
| Position | 4 | 5 | 6 | 7 | ||||
| δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | |
| 2 | 166.58 C | 164.12 C | 162.7 C | 163.75 C | ||||
| 3 | 89.81 CH | 5.14, s | 88.4 CH | 5.46 (d, 2.0) | 88.35 CH | 5.44 (d, 2.7) | 89.98 CH | 5.53 (d, 2.3) |
| 4 | 173.32 C | 171.11 C | 171.18 C | 170.56 C | ||||
| 5 | 29.55 CH2 | a. 2.97 (dd, 13.8, 17.3) b. 2.27 (dd, 3.9, 17.3) | 100.45 CH | 6.16 (d, 2.0) | 100.19 CH | 6.14 (d, 2.0) | 103.13 CH | 6.01, m |
| 6 | 75.94 CH | 4.51 (dt, 3.2, 3.2, 12.8) | 163.19 C | - | 167.3 C | - | 150.90 CH | 7.35 (d, 6.1) |
| 4-OCH3 | 56.18 | 3.76, s | 55.99 | 3.83, s | 55.96 | 3.82, s | 55.51 | 3.81, s |
| 1’ | 74.79 CH | 3.49 (dd, 2.0, 6.1) | 73.44 CH | 4.44 (d, 4.0) | 73.75 CH | 4.51, br,m | - | - |
| 2’ | 72.62 CH | 4.32 (t, 6.4) | 72.56 CH | 4.02, m | 72.98 CH | 4.51, br,m | - | - |
| 3’ | 129.24 CH | 5.53 (dd, 7.7, 15.7) | 33.24 CH2 | 1.43, m | 127.1 CH | 5.5 (dd, 1.3, 2.0, 15.5) | - | - |
| 4’ | 130.60 CH | 5.88, m | 18.91 CH2 | a. 1.54, m b. 1.35, m | 131.48 CH | 5.83 (dq, 6.7, 15.5) | - | - |
| 5’ | 17.88 CH3 | 1.74 (d, 6.4) | 13.91 CH3 | 0.93 (t, 7.4) | 17.9 CH3 | 1.71 (dd, 1.3, 6.7) | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, V.-T.; Bertrand, S.; Robiou du Pont, T.; Fleury, F.; Caroff, N.; Bourgeade-Delmas, S.; Gentil, E.; Logé, C.; Genta-Jouve, G.; Grovel, O. Untargeted Metabolomics Approach for the Discovery of Environment-Related Pyran-2-Ones Chemodiversity in a Marine-Sourced Penicillium restrictum. Mar. Drugs 2021, 19, 378. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070378
Le V-T, Bertrand S, Robiou du Pont T, Fleury F, Caroff N, Bourgeade-Delmas S, Gentil E, Logé C, Genta-Jouve G, Grovel O. Untargeted Metabolomics Approach for the Discovery of Environment-Related Pyran-2-Ones Chemodiversity in a Marine-Sourced Penicillium restrictum. Marine Drugs. 2021; 19(7):378. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070378
Chicago/Turabian StyleLe, Van-Tuyen, Samuel Bertrand, Thibaut Robiou du Pont, Fabrice Fleury, Nathalie Caroff, Sandra Bourgeade-Delmas, Emmanuel Gentil, Cedric Logé, Gregory Genta-Jouve, and Olivier Grovel. 2021. "Untargeted Metabolomics Approach for the Discovery of Environment-Related Pyran-2-Ones Chemodiversity in a Marine-Sourced Penicillium restrictum" Marine Drugs 19, no. 7: 378. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070378