Polymorphisms of HLA-DM on Treatment Response to Interferon/Ribavirin in Patients with Chronic Hepatitis C Virus Type 1 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Subjects
2.3. Laboratory Testing
2.4. SNP Genotyping
2.5. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Population
3.2. Association of Polymorphisms in HLA-DM with Virological Response to Treatment
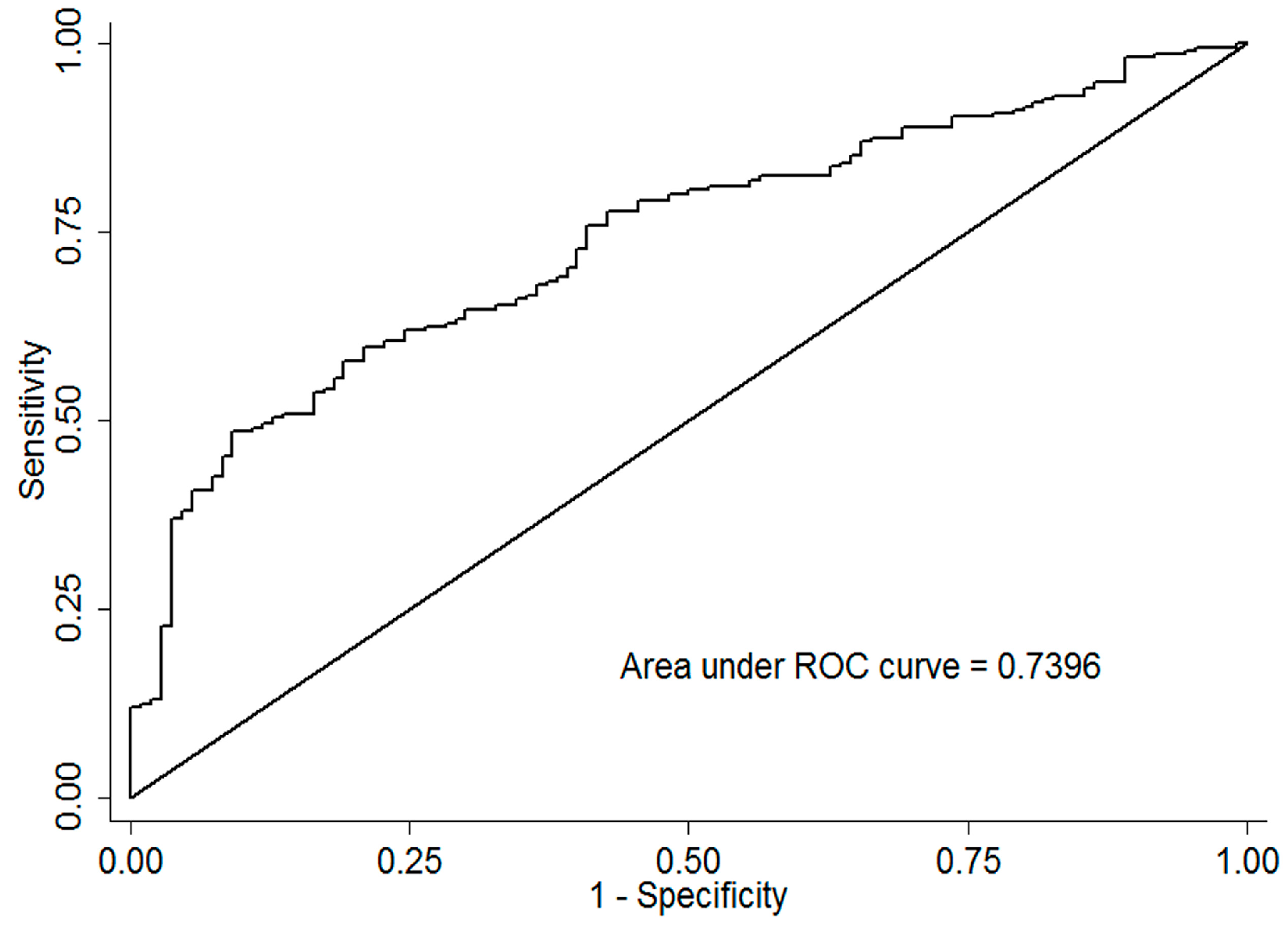
3.3. Predictive Factors for SVR
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lavanchy, D. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 2011, 17, 107–115. [Google Scholar] [PubMed]
- Alter, M.J. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 2007, 13, 2436. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, C.; Hézode, C.; Zeuzem, S.; Pawlotsky, J.-M. Antiviral strategies in hepatitis C virus infection. J. Hepatol. 2012, 56, S88–S100. [Google Scholar] [CrossRef]
- Nguyen, L.; Nguyen, M. Systematic review: Asian patients with chronic hepatitis C infection. Aliment. Pharmacol. Ther. 2013, 37, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Luo, J.; Bai, J.; Yu, R. Epidemiology of hepatitis C virus infection among injection drug users in China: Systematic review and meta-analysis. Public Health 2008, 122, 990–1003. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, F.; Hoang, T.; Kramer, J.R.; Asch, S.M.; Goetz, M.B.; Zeringue, A.; Richardson, P.; El–Serag, H.B. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C virus infection. Gastroenterology 2011, 140, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.M.; Stevens, C.R.; Walsh, E.C.; De Jager, P.L.; Goyette, P.; Plenge, R.M.; Vyse, T.J.; Rioux, J.D. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet. 2008, 4, e1000024. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Katsumata, K.; Atsumi, T.; Romero, F.; Bertolaccini, M.; Funke, A.; Amengual, O.; Kondeatis, E.; Vaughan, R.; Cox, A. Association of HLA-DM polymorphism with the production of antiphospholipid antibodies. Ann. Rheum. Dis. 2004, 63, 1645–1648. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.; Simoes, C.D.S.; Avinens, O.; Sany, J.; Combe, B.; Eliaou, J.-F. Polymorphism of HLA-DMA and DMB alleles in patients with systemic lupus erythematosus. J. Rheumatol. 2003, 30, 1485–1490. [Google Scholar] [PubMed]
- Perdriger, A.; Guggenbuhl, P.; Chales, G.; Yaouanq, J.; Quelvennec, E.; Bonnard, M.; Pawlotsky, Y.; Semana, G. Positive association of the HLA DMB1* 0101-0101 genotype with rheumatoid arthritis. Rheumatology 1999, 38, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Yee, L.J.; Im, K.; Wahed, A.S.; Bugawan, T.; Li, J.; Rhodes, S.L.; Erlich, H.; Rosen, H.R.; Liang, T.J.; Yang, H. Polymorphism in the human major histocompatibility complex and early viral decline during treatment of chronic hepatitis C. Antimicrob. Agents Chemother. 2009, 53, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Dong, L.; Lu, X.; Zhang, Y.; Chen, H.; Wang, J.; Zhang, Y.; Su, J.; Yu, R. Genetic variants in antigen presentation-related genes influence susceptibility to hepatitis C virus and viral clearance: A case control study. BMC Infect. Dis. 2014, 14, 716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Y.; Chen, H.-B.; Xu, Y.; Huang, P.; Wang, J.; Zhang, Y.; Yu, R.-B.; Su, J. Interferon-λ-related genes and therapeutic response in Chinese hepatitis C patients. World J. Gastroenterol. 2015, 21, 4006–4013. [Google Scholar] [CrossRef] [PubMed]
- Ochi, H.; Hayes, C.N.; Abe, H.; Hayashida, Y.; Uchiyama, T.; Kamatani, N.; Nakamura, Y.; Chayama, K. Toward the establishment of a prediction system for the personalized treatment of chronic hepatitis C. J. Infect. Dis. 2012, 205, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, P.; Yue, M.; Su, J.; Chen, H.; Chen, M.; Wang, J.; Li, J.; Peng, Z.; Zhang, Y. A novel polymorphism near HLA class II region is associated with spontaneous clearance of HCV and response to interferon treatment in Chinese patients. J. Hum. Genet. 2016, 61, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; He, X. Association of HLA-DM gene with childhood systemic lupus erythematosus. Natl. Med. J. China 2013, 93, 984–986. (In Chinese) [Google Scholar]
- Aissani, B.; Boehme, A.K.; Wiener, H.W.; Shrestha, S.; Jacobson, L.P.; Kaslow, R.A. SNP screening of central MHC-identified HLA-DMB as a candidate susceptibility gene for HIV-related Kaposi’s sarcoma. Genes Immun. 2014, 15, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Andriulli, A.; Di Marco, V.; Margaglione, M.; Ippolito, A.M.; Fattovich, G.; Smedile, A.; Valvano, M.R.; Calvaruso, V.; Gioffreda, D.; Milella, M. Identification of naive HCV-1 patients with chronic hepatitis who may benefit from dual therapy with peg-interferon and ribavirin. J. Hepatol. 2014, 60, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Variables | N-SVR | SVR | p-Value |
|---|---|---|---|
| (n = 83) | (n = 253) | ||
| Mean age, years | 53.60 ± 8.05 | 53.52 ± 8.57 | 0.917 |
| Age ≥ 50 | 77 (69.4) | 151 (67.1) | 0.677 |
| Male (%) | 27 (24.3) | 56 (24.9) | 0.910 |
| AST ≥ 40 U/L (%) | 61 (54.9) | 109 (48.4) | 0.262 |
| ALT ≥ 40 U/L (%) | 67 (63.4) | 125 (55.6) | 0.403 |
| GGT ≥ 50 (U/L) | 50 (45.1) | 71 (31.6) | 0.015 |
| GLU > 6 (mmol/L) | 49 (44.1) | 58 (25.8) | 0.001 |
| AFP > 7.02 (ng/mL) | 36 (32.4) | 45 (20.0) | 0.012 |
| T3 (nmol/L) | 1.55 ± 0.44 | 1.41 ± 0.45 | 0.008 |
| T4 (nmol/L) | 130.94 ± 33.45 | 121.55 ± 30.48 | 0.011 |
| Anti-TPO ≥ 35 I/mL | 16 (14.5) | 33 (15.0) | 0.913 |
| baseline HCV-RNA | 6.21 ± 0.74 | 6.00 ± 0.85 | 0.032 |
| TP (g/L) | 79.79 ± 5.84 | 78.84 ± 6.80 | 0.061 |
| ALB (g/L) | 43.34 ± 4.25 | 43.84 ± 4.29 | 0.316 |
| Platelets (109/L) | 133.47 ± 68.85 | 147.48 ± 60.63 | 0.058 |
| Abnormal | 47 (42.3) | 58 (25.9) | 0.002 |
| Normal | 64 (57.7) | 166 (74.1) | |
| WBC (109/L) | 4.79 ± 1.79 | 4.96 ± 1.72 | 0.394 |
| Abnormal | 43 (39.1) | 72 (32.0) | 0.199 |
| Normal | 67 (60.9) | 153 (68.0) | |
| Hemoglobin (g/L) | 133.52 ± 17.86 | 132.94 ± 17.55 | 0.782 |
| Abnormal | 31 (27.9) | 56 (24.9) | 0.550 |
| Normal | 80 (72.1) | 169 (75.1) |
| Genotype | N-SVR | SVR | SVR Rate (%) | OR (95% CI) | p-Value |
|---|---|---|---|---|---|
| rs23544 | |||||
| AA | 62 (55.9) | 80 (35.6) | 56.3 | 1.00 | - |
| AG | 40 (36.0) | 100 (44.4) | 71.4 | 1.74 (1.02–2.98) | 0.044 |
| GG | 9 (8.1) | 45 (20.0) | 83.3 | 3.22 (1.41–7.35) | 0.006 |
| Dominant | 2.04 (1.23–3.35) | 0.005 | |||
| Recessive | 2.52 (1.14–5.58) | 0.022 | |||
| Additive | 1.78 (1.23–2.56) | 0.002 | |||
| rs3135029 | |||||
| AA | 71 (64.0) | 155 (68.9) | 68.6 | 1.00 | - |
| AC | 32 (28.8) | 63 (28.0) | 66.3 | 1.04 (0.59–1.83) | 0.879 |
| CC | 8 (7.2) | 7 (3.1) | 46.7 | 0.37 (0.11–1.19) | 0.097 |
| Dominant | 0.89 (0.53–1.51) | 0.686 | |||
| Recessive | 0.36 (0.11–1.17) | 0.090 | |||
| Additive | 0.81 (0.52–1.24) | 0.337 | |||
| rs1050391 | |||||
| CC | 72 (64.9) | 156 (69.3) | 68.4 | 1.00 | - |
| CT | 33 (29.7) | 62 (27.6) | 65.3 | 0.96 (0.55–1.69) | 0.906 |
| TT | 6 (5.4) | 7 (3.1) | 53.8 | 0.40 (0.12–1.37) | 0.148 |
| Dominant | 0.86 (0.51–1.46) | 0.577 | |||
| Recessive | 0.41 (0.12–1.37) | 0.149 | |||
| Additive | 0.80 (0.52–1.24) | 0.331 | |||
| rs1063478 | |||||
| CC | 59 (53.2) | 86 (38.2) | 59.3 | 1.00 | - |
| CT | 46 (41.4) | 111 (49.3) | 70.7 | 1.83 (1.08–3.08) | 0.022 |
| TT | 6 (5.4) | 28 (12.5) | 82.3 | 4.40 (1.48–13.03) | 0.007 |
| Dominant | 2.05 (1.24–3.41) | 0.005 | |||
| Recessive | 3.15 (1.11–8.96) | 0.031 | |||
| Additive | 1.96 (1.29–2.96) | 0.002 |
| Variables | N-SVR | SVR | SVR Rate (%) | OR (95% CI) | p-Value |
|---|---|---|---|---|---|
| 0 | 28 (25.2) | 22 (9.8) | 44.0 | 1.00 | - |
| 1 | 51 (46.0) | 94 (41.8) | 64.8 | 2.27 (1.10–4.63) | 0.025 |
| 2 | 31 (27.9) | 74 (32.9) | 70.5 | 2.91 (1.37–6.17) | 0.005 |
| 3–4 | 1 (0.9) | 35 (15.5) | 97.2 | 45.12 (5.49–370.75) | <0.001 |
| Trend | a p < 0.001 |
| Variables | Coef. | SE | OR (95% CI) | p-Value |
|---|---|---|---|---|
| rs23544 | 0.68 | 0.37 | 1.98 (1.36–2.89) | <0.001 |
| rs1063478 | 0.79 | 0.47 | 2.22 (1.45–3.38) | <0.001 |
| GLU | −0.77 | 0.12 | 0.46 (0.27–0.78) | 0.004 |
| Platelets (109/L) | 0.70 | 0.52 | 2.02 (1.21–3.34) | 0.007 |
| T4 | −0.008 | 0.004 | 0.99 (0.98–1.00) | 0.046 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Yao, Y.; Wang, Y.; Zhou, H.; Xu, T.; Liu, J.; Wang, G.; Zhang, Y.; Chen, X.; Liu, Q.; et al. Polymorphisms of HLA-DM on Treatment Response to Interferon/Ribavirin in Patients with Chronic Hepatitis C Virus Type 1 Infection. Int. J. Environ. Res. Public Health 2016, 13, 1030. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13101030
Chen H, Yao Y, Wang Y, Zhou H, Xu T, Liu J, Wang G, Zhang Y, Chen X, Liu Q, et al. Polymorphisms of HLA-DM on Treatment Response to Interferon/Ribavirin in Patients with Chronic Hepatitis C Virus Type 1 Infection. International Journal of Environmental Research and Public Health. 2016; 13(10):1030. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13101030
Chicago/Turabian StyleChen, Hongbo, Yinan Yao, Yifan Wang, Hua Zhou, Tianxiang Xu, Jing Liu, Guocheng Wang, Yongfeng Zhang, Xiang Chen, Qingwei Liu, and et al. 2016. "Polymorphisms of HLA-DM on Treatment Response to Interferon/Ribavirin in Patients with Chronic Hepatitis C Virus Type 1 Infection" International Journal of Environmental Research and Public Health 13, no. 10: 1030. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13101030





