Bacterial Biotransformation of Pentachlorophenol and Micropollutants Formed during Its Production Process
Abstract
:1. Introduction
2. Toxicity and Bacterial Degradation of PCP
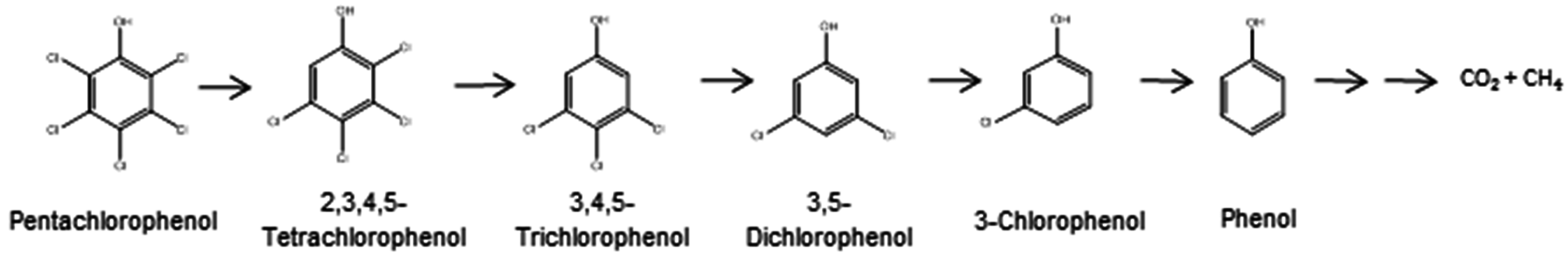
2.1. Aerobic Bacterial Degradation of PCP
2.2. Anaerobic Reductive Dechlorination of PCP
3. Biodegradation of Polychlorinated Dibenzo-p-Dioxins (PCDDs) and Polychlorinated Dibenzofurans (PCDFs)
3.1. PCDD/Fs Toxicity
3.2. Biodegradation of PCDD/Fs
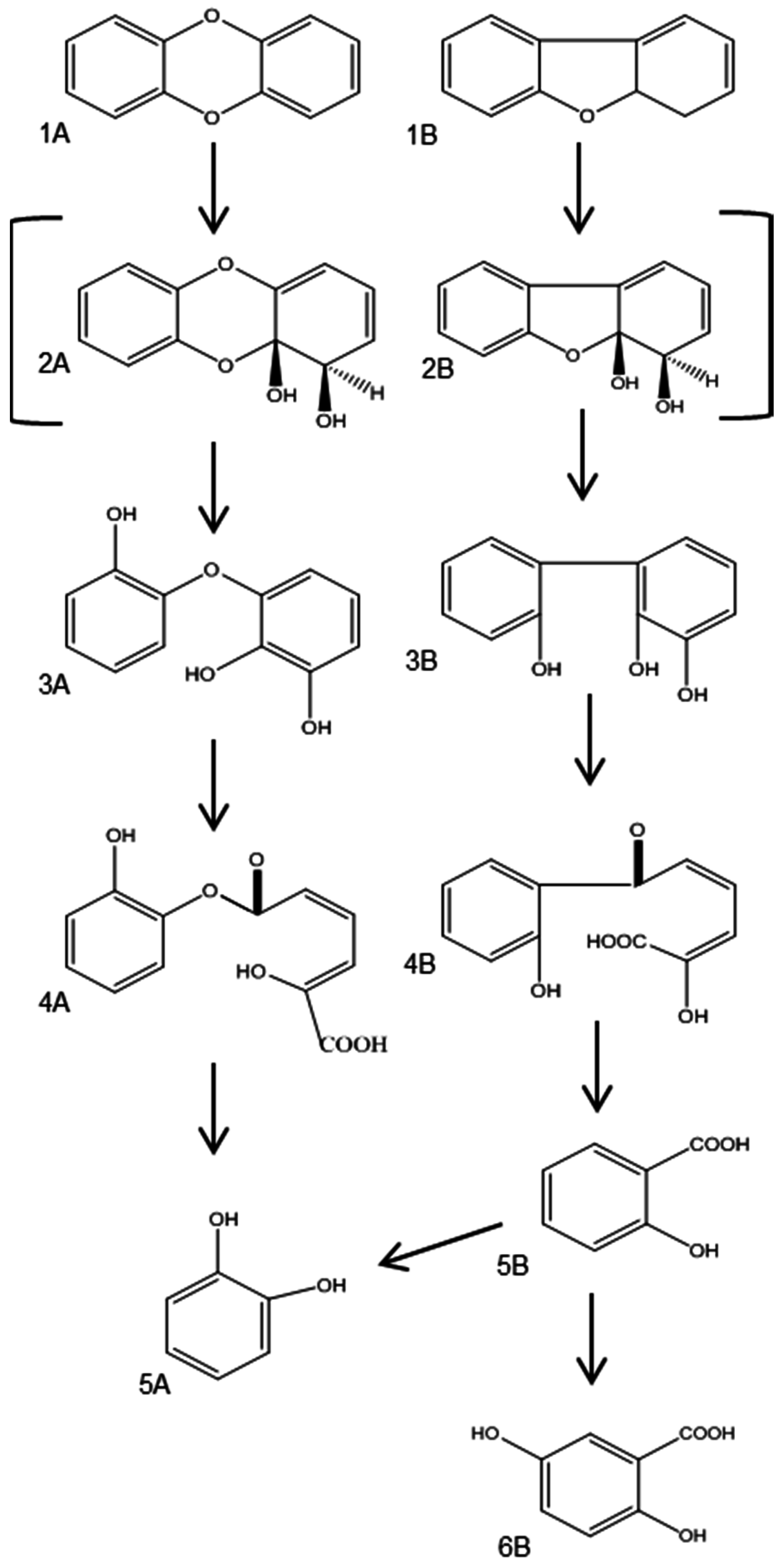
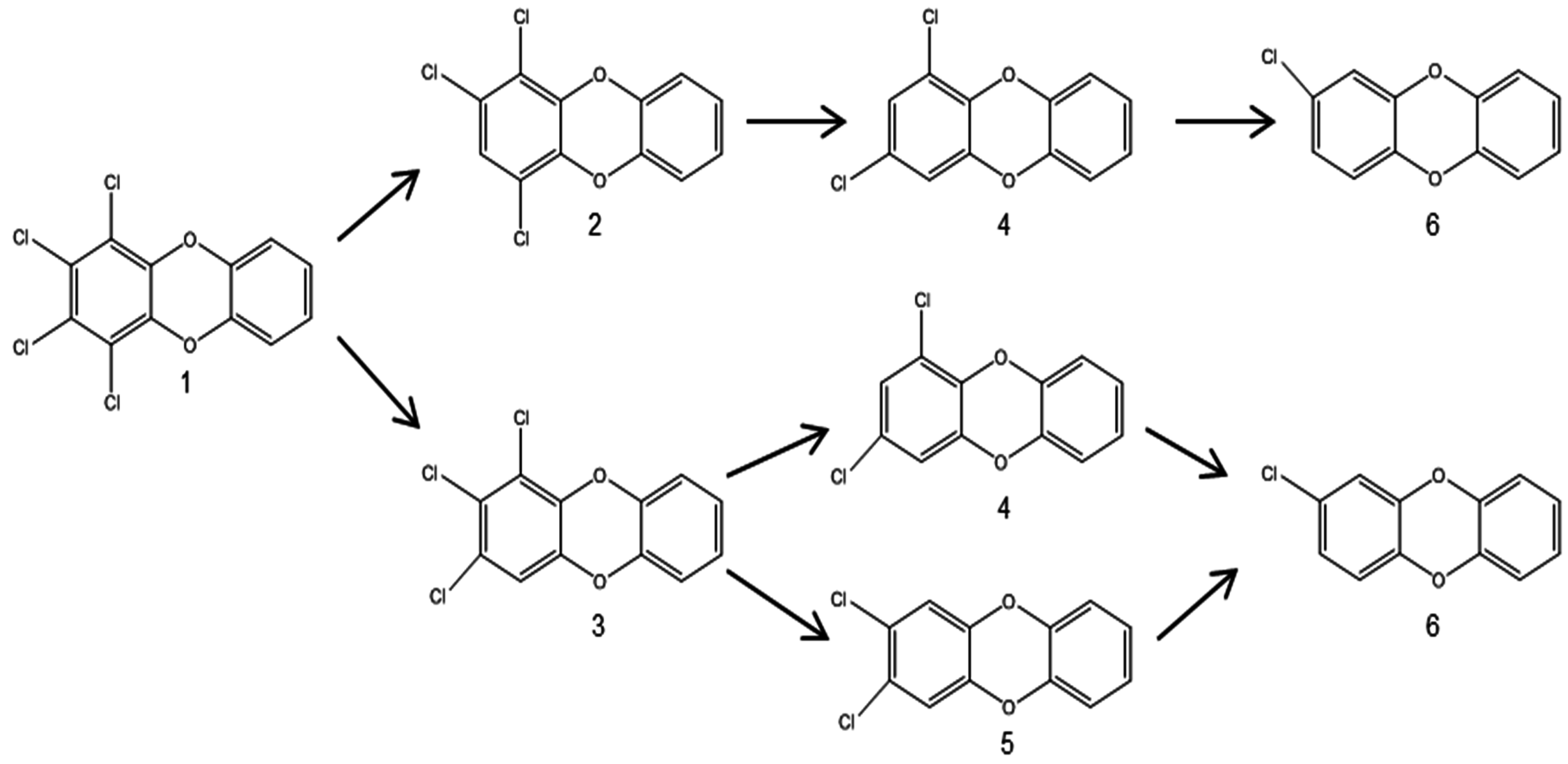
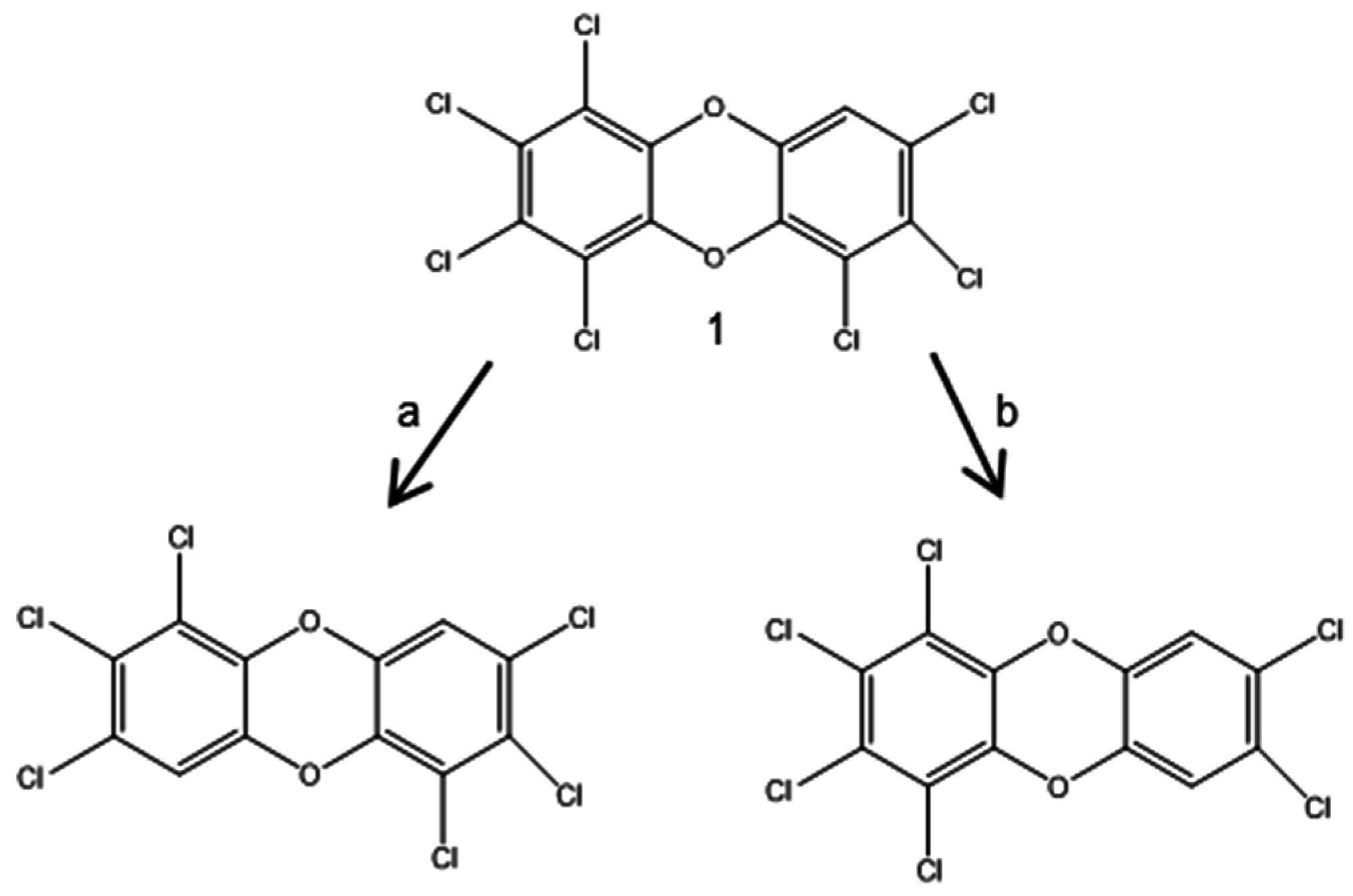
3.2.1. Aerobic Bacterial Degradation of PCDD/Fs
3.2.2. Anaerobic Reductive Dechlorination of PCDD/Fs
4. Bioremediation Studies
4.1. Composting and Soil Biopiles
4.2. Bioreactors and Microcosms
4.3. Phytoremediation
5. Microbial Communities in Contaminated Environments
5.1. Bacterial Communities In Situ
5.2. Bacterial Communities in Bioremediation Studies
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AHD | aromatic-hydrocarbon-degrading |
| AI | active ingredient |
| CMC | carboxymethylcellulose |
| CDD/Fs | chlorinated dibenzo-p-dioxins and dibenzofurans |
| CNP | 2,4,6-trichlorophenyl-4′-nitrophenyl ether |
| CP | chlorophenol |
| CPs | chlorinated phenols |
| d.w. | dry weight |
| DCP | dichlorophenol |
| DCDD | dichlorinated dibenzo-p-dioxin |
| DCDF | dichlorinated dibenzofuran |
| DD | dibenzo-p-dioxin |
| DF | dibenzofuran |
| FBC | fed-batch composting |
| HeCDD | heptachlorinated dibenzo-p-dioxin |
| HeCDF | heptachlorinated dibenzofuran |
| HxCDD | hexachlorinated dibenzo-p-dioxin |
| HxCDF | hexachlorinated dibenzofuran |
| MCPs | monochlorinated phenols |
| MSW | municipal solid waste |
| OCDD | octachlorinated dibenzo-p-dioxin |
| OCDF | octachlorinated dibenzofuran |
| PCDD | polychlorinated dibenzo-p-dioxin |
| PCP | pentachlorophenol |
| PeCDD | pentachlorinated dibenzo-p-dioxin |
| PCDF | polychlorinated dibenzofuran |
| PeCDF | pentachlorinated dibenzofuran |
| RDases | reductive dehalogenases |
| TCA | tricarboxylic acid |
| TCDD | trichlorinated dibenzo-p-dioxin |
| TCDF | trichlorinated dibenzofuran |
| TCHQ | tetrachlorohydroquinone |
| TCP | trichlorophenol |
| TeCDD | tetrachlorinated dibenzo-p-dioxin |
| TeCDF | tetrachlorinated dibenzofuran |
| TeCP | tetrachlorophenol |
| TEQ | toxic equivalent |
References
- World Health Organization (WHO). Pentachlorophenol. Available online: http://www.inchem.org/documents/ehc/ehc/ehc71.htm (accessed on 17 July 2015).
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Pentachlorophenol; Update; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 2001.
- EuroChlor. Pentachlorophenol; Euro Chlor Risk Assessment for the Marine Environment; OSPARCOM Region—North Sea; EuroChlor: Brussels, Belgium, 1999. [Google Scholar]
- World Health Organization (WHO). Pentachlorophenol in Drinking-Water; Background Document for Development of WHO Guidelines for Drinking-Water Quality; World Health Organization: Geneva, Switzerland, 1998. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO). Pentachlorophenol and Its Salts and Esters; Decision Guidance Documents; United Nations Environment Programme: Rome, Italy; Geneva, Switzerland, 1996. [Google Scholar]
- Muller, F.; Caillard, L. Chlorophenols. Ullmann’s Encycl. Ind. Chem. 2011, 45, 43–50. [Google Scholar]
- Fisher, B. Pentachlorophenol: Toxicology and environmental fate. J. Pestic. Reform 1991, 11, 1–5. [Google Scholar]
- Yu, J.; Nestrick, T.J.; Allen, R.; Savage, P.E. Microcontaminants in pentachlorophenol synthesis. 1. New bioassay for microcontaminant quantification. Ind. Eng. Chem. Res. 2006, 45, 5199–5204. [Google Scholar] [CrossRef]
- Holt, E.; Weber, R.; Stevenson, G.; Gaus, C. Polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs) impurities in pesticides: A neglected source of contemporary relevance. Environ. Sci. Technol. 2010, 44, 5409–5415. [Google Scholar] [CrossRef] [PubMed]
- Anasonye, F.; Winquist, E.; Kluczek-Turpeinen, B.; Rasanen, M.; Salonen, K.; Steffen, K.T.; Tuomela, M. Fungal enzyme production and biodegradation of polychlorinated dibenzo-p-dioxins and dibenzofurans in contaminated sawmill soil. Chemosphere 2014, 110, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Persson, Y.; Lundstedt, S.; Oberg, L.; Tysklind, M. Levels of chlorinated compounds (CPs, PCPPs, PCDEs, PCDFs and PCDDs) in soils at contaminated sawmill sites in Sweden. Chemosphere 2007, 66, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Kitunen, V.H.; Valo, R.J.; Salkinoja-Salonen, M.S. Contamination of soil around wood-preserving facilities by polychlorinated aromatic compounds. Environ. Sci. Technol. 1987, 21, 96–101. [Google Scholar] [CrossRef]
- Salo, S.; Verta, M.; Malve, O.; Korhonen, M.; Lehtoranta, J.; Kiviranta, H.; Isosaari, P.; Ruokojarvi, P.; Koistinen, J.; Vartiainen, T. Contamination of River Kymijoki sediments with polychlorinated dibenzo-p-dioxins, dibenzofurans and mercury and their transport to the Gulf of Finland in the Baltic Sea. Chemosphere 2008, 73, 1675–1683. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; Masunaga, S. PCDD/PCDF contamination from historical pesticide use and production—A case study using data from Japan and Germany. In Proceedings of the International HCH and Pesticides Forum, Sofia, Bulgaria, 26–28 May 2005.
- Nie, Z.; Tang, Z.; Zhu, X.; Yang, Y.; Fu, H.; Die, Q.; Wang, Q.; Huang, Q. Occurrence, possible sources, and temporal trends of polychlorinated dibenzo-p-dioxins and dibenzofurans in water and sediment from the lower Yangtze River basin, Jiangsu and Shanghai areas of Eastern China. Environ. Sci. Pollut. Res. 2013, 20, 8751–8762. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Xu, Y.; Brookes, P.C.; He, Y.; Xu, J. Spatial and temporal variations in pentachlorophenol dissipation at the aerobic—Anaerobic interfaces of flooded paddy soils. Environ. Pollut. 2013, 178, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.K.; Bae, H. Bacterial degradation of chlorophenols and their derivatives. Microb. Cell Fact. 2014, 13. [Google Scholar] [CrossRef] [PubMed]
- Orser, C.; Lange, C. Molecular analysis of pentachlorophenol degradation. Biodegradation 1994, 5, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Kozak, V.P.; Simsiman, G.V.; Chesters, G.; Stensby, D.; Harkin, J. Reviews of the Environmental Effects of Pollutants. XI. Chlorophenols; U.S. Environmental Protection Agency: Washington, DC, USA, 1979; p. 492.
- U.S. Environmental Protection Agency (EPA). Integrated Risk Information System (IRIS) on Pentachlorophenol. Available online: http://www.epa.gov/iris/subst/0086.htm (accessed on 14 May 2015).
- Cooper, G.S.; Jones, S. Pentachlorophenol and cancer risk: Focusing the lens on specific chlorophenols and contaminants. Environ. Health Perspect. 2008, 116, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Hamana, K.; Hiraishi, A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Bacterial dehalogenation. Appl. Microbiol. Biotechnol. 1998, 50, 633–657. [Google Scholar] [CrossRef] [PubMed]
- Xun, L.; Orser, C.S. Purification and properties of pentachlorophenol hydroxylase, a flavoprotein from Flavobacterium sp. strain ATCC 39723. J. Bacteriol. 1991, 173, 4447–4453. [Google Scholar] [CrossRef] [PubMed]
- Orser, C.S.; Lange, C.C.; Xun, L.; Zahrt, T.C.; Schneider, B.J. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 1993, 175, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.C.; Schneider, B.J.; Orser, C.S. Verification of the role of pcp 4-monooxygenase in chlorine elimination from pentachlorophenol by Flavobacterium sp. strain ATCC 39723. Biochem. Biophys. Res. Commun. 1996, 219, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Orser, C.S.; Dutton, J.; Lange, C.; Jablonski, P.; Xun, L.; Hargis, M. Characterization of a Flavobacterium glutathione S-transferase gene involved reductive dechlorination. J. Bacteriol. 1993, 175, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, Y.; Miyauchi, K.; Kanda, K.; Hatta, T.; Kiyohara, H.; Senda, T.; Nagata, Y.; Mitsui, Y.; Takagi, M. PcpA, which is involved in the degradation of pentachlorophenol in Sphingomonas chlorophenolica ATCC39723, is a novel type of ring-cleavage dioxygenase. FEBS Lett. 1999, 459, 395–398. [Google Scholar] [CrossRef]
- Steiert, J.G.; Crawford, R.L. Catabolism of pentachlorophenol by a Flavobacterium sp. Biochem. Biophys. Res. Commun. 1986, 141, 825–830. [Google Scholar] [CrossRef]
- Uotila, J.S.; Kitunen, V.H.; Saastamoinen, T.; Coote, T.; Häggblom, M.M.; Salkinoja-Salonen, M.S. Characterization of Aromatic Dehalogenases of Mycobacterium fortuitum CG-2. J. Bacteriol. 1992, 174, 5669–5675. [Google Scholar] [CrossRef] [PubMed]
- Uotila, J.S.; Salkinoja-Salonen, M.S.; Apajalahti, J.H. Dechlorination of pentachlorophenol by membrane bound enzymes of Rhodococcus chlorophenolicus PCP-I. Biodegradation 1991, 2, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Apajalahti, J.H.A.; Salkinoja-Salonen, M.S. Complete dechlorination of tetrachlorohydroquinone by cell extracts of pentachlorophenol-induced Rhodococcus chlorophenolicus. J. Bacteriol. 1987, 169, 5125–5130. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A. Biodiversity of dehalorespiring bacteria with special emphasis on polychlorinated biphenyl/dioxin dechlorinators. Microbes Environ. 2008, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Field, J.A.; Sierra-Alvarez, R. Microbial degradation of chlorinated phenols. Rev. Environ. Sci. Bio/Technol. 2008, 7, 211–241. [Google Scholar] [CrossRef]
- Holliger, C.; Wohlfarth, G.; Diekert, G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 1998, 22, 383–398. [Google Scholar] [CrossRef]
- Bunge, M.; Lechner, U. Anaerobic reductive dehalogenation of polychlorinated dioxins. Appl. Microbiol. Biotechnol. 2009, 84, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Jugder, B.-E.; Ertan, H.; Bohl, S.; Lee, M.; Marquis, C.P.; Manefield, M. Organohalide respiring bacteria and reductive dehalogenases: Key tools in organohalide bioremediation. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, P.; Grbic’-Galic, D. Reductive dechlorination of PCDD/F by anaerobic cultures and sediments. Chemosphere 1994, 29, 2253–2259. [Google Scholar] [CrossRef]
- Richardson, R.E. Genomic insights into organohalide respiration. Curr. Opin. Biotechnol. 2013, 24, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Mikesell, M.D.; Boyd, S.A. Complete reductive dechlorination and mineralization of pentachlorophenol by anaerobic microorganisms. Appl. Environ. Microbiol. 1986, 52, 861–865. [Google Scholar] [PubMed]
- Stuart, S.L.; Woods, S.L. Kinetic evidence for pentachlorophenol-dependent growth of a dehalogenating population in a pentachlorophenol- and acetate-fed methanogenic culture. Biotechnol. Bioeng. 1998, 57, 420–429. [Google Scholar] [CrossRef]
- Villemur, R. The pentachlorophenol-dehalogenating Desulfitobacterium hafniense strain PCP-1. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef] [PubMed]
- Van de Pas, B.A.; Smidt, H.; Hagen, W.R.; van der Oost, J.; Schraa, G.; Stams, A.J.M.; de Vos, W.M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 1999, 274, 20287–20292. [Google Scholar] [CrossRef] [PubMed]
- Bisaillon, A.; Beaudet, R.; Lepine, F.; Deziel, E.; Villemur, R. Identification and characterization of a novel CprA reductive dehalogenase specific to highly chlorinated phenols from Desulfitobacterium hafniense strain PCP-1. Appl. Environ. Microbiol. 2010, 76, 7536–7540. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, J.; Gauthier, A.; Duguay, M.; Villemur, R.; Lepine, F.; Juteau, P.; Beaudet, R. Purification, cloning, and sequencing of a 3,5-dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 2004, 70, 4532–4537. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.; Page-BeLanger, R.; Saucier, M.; Villemur, R.; Lepine, F.; Juteau, P.; Beaudet, R. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6-trichlorophenol from Desulfitobacterium frappieri PCP-1. Biochem. J. 2003, 373, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Chlorinated Dibenzo-p-Dioxins (CDDs). Available online: http://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=366&tid=63 (accessed on 9 July 2015).
- Barring, H.; Bucheli, T.D.; Broman, D.; Gustafsson, O. Soot-water distribution coefficients for polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans and polybrominated diphenylethers determined with the soot cosolvency-column method. Chemosphere 2002, 49, 515–523. [Google Scholar] [CrossRef]
- Cornelissen, G.; Gustafsson, Ö.; Bucheli, T.D.; Jonker, M.T.; Koelmans, A.A.; van Noort, P.C. Extensive sorption of organic compounds to black carbon, coal, and kerogen in sediments and soils: Mechanisms and consequences for distribution, bioaccumulation, and biodegradation. Environ. Sci. Technol. 2005, 39, 6881–6895. [Google Scholar] [CrossRef] [PubMed]
- Srogi, K. Levels and congener distributions of PCDDs, PCDFs and dioxin-like PCBs in environmental and human samples: A review. Environ. Chem. Lett. 2008, 6, 1–28. [Google Scholar] [CrossRef]
- Otles, S.; Yildiz, H. Dioxin in food and human health. Electron. J. Environ. Agric. Food Chem. 2003, 2, 593–608. [Google Scholar]
- Hoekstra, E.J.; de Weerd, H.; de Leer, E.W.; Brinkman, U.A.T. Natural formation of chlorinated phenols, dibenzo-p-dioxins, and dibenzofurans in soil of a Douglas fir forest. Environ. Sci. Technol. 1999, 33, 2543–2549. [Google Scholar] [CrossRef]
- Lavric, E.D.; Konnov, A.A.; De Ruyck, J. Dioxin levels in wood combustion—A review. Biomass Bioenergy 2004, 26, 115–145. [Google Scholar] [CrossRef]
- Gu, C.; Li, H.; Teppen, B.J.; Boyd, S.A. Octachlorodibenzodioxin formation on Fe(III)-montmorillonite clay. Environ. Sci. Technol. 2008, 42, 4758–4763. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, C.; Ding, Y.; Li, H.; Teppen, B.J.; Johnston, C.T.; Boyd, S.A. Clay mediated route to natural formation of Polychlorodibenzo-p-dioxins. Environ. Sci. Technol. 2011, 45, 3445–3451. [Google Scholar] [CrossRef] [PubMed]
- Wittich, R.-M. Degradation of dioxin-like compounds by microorganisms. Appl. Microbiol. Biotechnol. 1998, 49, 489–499. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Dioxins and Their Effects on Human Health. Available online: http://www.who.int/mediacentre/factsheets/fs225/en/ (accessed on 19 July 2015).
- World Health Organization (WHO). Polychlorinated Dibenzo-p-dioxins and Dibenzofurans. Available online: http://www.inchem.org/documents/ehc/ehc/ehc88.htm (accessed on 23 June 2015).
- Dahlgren, J.; Warshaw, R.; Horsak, R.D.; Parker, F.M., III; Takhar, H. Exposure assessment of residents living near a wood treatment plant. Environ. Res. 2003, 92, 99–109. [Google Scholar] [CrossRef]
- Fries, G.F.; Feil, V.J.; Zaylskie, R.G.; Bialek, K.M.; Rice, C.P. Treated wood in livestock facilities: Relationships among residues of pentachlorophenol, dioxins, and furans in wood and beef. Environ. Pollut. 2002, 116, 301–307. [Google Scholar] [CrossRef]
- Huwe, J.K.; Davison, K.; Feil, V.J.; Larsen, G.; Lorentzsen, M.; Zaylskie, R.; Tiernan, T.O. Levels of polychlorinated dibenzo-p-dioxins and dibenzofurans in cattle raised at agricultural research facilities across the USA and the influence of pentachlorophenol-treated wood. Food Addit.Contam. 2004, 21, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Field, J.A.; Sierra-Alvarez, R. Microbial degradation of chlorinated dioxins. Chemosphere 2008, 71, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.B.; Nam, I.H.; Murugesan, K.; Kim, Y.M.; Chang, Y.S. Biodegradation of dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins by Pseudomonas veronii PH-03. Biodegradation 2004, 15, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zanaroli, G.; Negroni, A.; Häggblom, M.M.; Fava, F. Microbial dehalogenation of organohalides in marine and estuarine environments. Curr. Opin. Biotechnol. 2015, 33, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A. Biodiversity of dioxin-degrading microorganisms and potential utilization in bioremediation. Microbes Environ. 2003, 18, 105–125. [Google Scholar] [CrossRef]
- Chang, Y.-S. Recent developments in microbial biotransformation and biodegradation of dioxins. J. Mol. Microbiol. Biotechnol. 2008, 15, 152–171. [Google Scholar] [CrossRef] [PubMed]
- Klečka, G.M.; Gibson, D.T. Metabolism of dibenzo-p-dioxin by a Pseudomonas species. Biochem. J. 1979, 180, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Wittich, R.M.; Wilkes, H.; Sinnwell, V.; Francke, W.; Fortnagel, P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 1992, 58, 1005–1010. [Google Scholar] [PubMed]
- Nojiri, H.; Omori, T. Molecular bases of aerobic bacterial degradation of dioxins: Involvement of angular dioxygenation. Biosci. Biotechnol. Biochem. 2002, 66, 2001–2016. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhu, T.; Xu, X.; Xu, Y. Biodegradation of dibenzofuran by Janibacter terrae strain XJ-1. Curr. Microbiol. 2006, 53, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Sukda, P.; Nishida, T.; Ito, E.; Matsumoto, Y.; Masai, E.; Fukuda, M. Isolation of dibenzofuran-degrading bacterium, Nocardioides sp. DF412, and characterization of its dibenzofuran degradation genes. J. Biosci. Bioeng. 2008, 105, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Sukda, P.; Gouda, N.; Ito, E.; Miyauchi, K.; Masai, E.; Fukuda, M. Characterization of a transcriptional regulatory gene involved in dibenzofuran degradation by Nocardioides sp. strain DF412. Biosci. Biotechnol. Biochem. 2009, 73, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, P.K.; Thakur, I.S. Isolation and characterization of dibenzofuran-degrading Serratia marcescens from alkalophilic bacterial consortium of the chemostat. Curr. Microbiol. 2007, 55, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, E.; Yamamoto, H.; Terakubo, S.; Okamura, N.; Naka, T.; Fujiwara, N.; Kobayashi, K.; Kosako, Y.; Hiraishi, A. Proposal of Sphingomonas wittichii sp. nov. for strain RW1T, known as a dibenzo-p-dioxin metabolizer. Int. J. Syst. Evol. Microbiol. 2001, 51, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Happe, B.; Timmis, K.N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: Catabolic genes dispersed on the genome. J. Bacteriol. 1998, 180, 3954–3966. [Google Scholar] [PubMed]
- Armengaud, J.; Timmis, K.N.; Wittich, R.M. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 1999, 181, 3452–3461. [Google Scholar] [PubMed]
- Wilkes, H.; Wittich, R.; Timmis, K.N.; Fortnagel, P.; Francke, W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 1996, 62, 367–371. [Google Scholar] [PubMed]
- Nam, I.H.; Kim, Y.M.; Schmidt, S.; Chang, Y.S. Biotransformation of 1,2,3-tri- and 1,2,3,4,7,8-hexachlorodibenzo-p-dioxin by Sphingomonas wittichii strain RW1. Appl. Environ. Microbiol. 2006, 72, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Ballerstedt, H.; Kraus, A.; Lechner, U. Reductive dechlorination of 1,2,3,4-tetrachlorodibenzo-p-dioxin and its products by anaerobic mixed cultures from Saale River sediment. Environ. Sci. Technol. 1997, 31, 1749–1753. [Google Scholar] [CrossRef]
- Ahn, Y.B.; Liu, F.; Fennell, D.E.; Haggblom, M.M. Biostimulation and bioaugmentation to enhance dechlorination of polychlorinated dibenzo-p-dioxins in contaminated sediments. FEMS Microbiol. Ecol. 2008, 66, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Beurskens, J.E.; De Wolf, J.; Toussaint, E.; van der Steen, J.; Slot, P.C.; Commandeur, L.; Parsons, J.R. Dehalogenation of chlorinated dioxins by an anaerobic microbial consortium from sediment. Environ. Toxicol. Chem. 1995, 14, 939–943. [Google Scholar] [CrossRef]
- Bunge, M.; Ballerstedt, H.; Lechner, U. Regiospecific dechlorination of spiked tetra- and trichlorodibenzo-p-dioxins by anaerobic bacteria from PCDD/F-contaminated Spittelwasser sediments. Chemosphere 2001, 43, 675–681. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Behrens, S.F.; Muller, J.A.; Goke, J.; Ritalahti, K.M.; Wagner, R.; Goltsman, E.; Lapidus, A.; Holmes, S.; Loffler, F.E.; et al. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009, 5, e1000714. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Segler, L.; Kleinsteuber, S.; Sawers, G.; Smidt, H.; Lechner, U. Regulation of reductive dehalogenase gene transcription in Dehalococcoides mccartyi. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120317. [Google Scholar] [CrossRef] [PubMed]
- Smidt, H.; de Vos, W.M. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 2004, 58, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63, 625–635. [Google Scholar] [PubMed]
- Seshadri, R.; Adrian, L.; Fouts, D.E.; Eisen, J.A.; Phillippy, A.M.; Methe, B.A.; Ward, N.L.; Nelson, W.C.; Deboy, R.T.; Khouri, H.M.; et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 2005, 307, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Beck, A.; Zinder, S.H.; Kuhl, H.; Reinhardt, R.; Adrian, L. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 2005, 23, 1269–1273. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chng, K.R.; Wilm, A.; Zhao, S.; Yang, K.-L.; Nagarajan, N.; He, J. Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc. Natl. Acad. Sci. USA 2014, 111, 12103–12108. [Google Scholar] [CrossRef] [PubMed]
- Laine, M.M.; Jørgensen, K.S. Straw compost and bioremediated soil as inocula for the bioremediation of chlorophenol-contaminated soil. Appl. Environ. Microbiol. 1996, 62, 1507–1513. [Google Scholar] [PubMed]
- Laine, M.M.; Jørgensen, K.S. Effective and safe composting of chlorophenol-contaminated soil in pilot scale. Environ. Sci. Technol. 1997, 31, 371–378. [Google Scholar] [CrossRef]
- Jaspers, C.J.; Ewbank, G.; McCarthy, A.J.; Penninckx, M.J. Successive rapid reductive dehalogenation and mineralization of pentachlorophenol by the indigenous microflora of farmyard manure compost. J. Appl. Microbiol. 2002, 92, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Yu, Z.; Chen, Y.; Zhang, J.; Li, H.; Yu, M.; Zhao, M. Response of compost maturity and microbial community composition to pentachlorophenol (PCP)-contaminated soil during composting. Bioresour. Technol. 2011, 102, 5905–5911. [Google Scholar] [CrossRef] [PubMed]
- Laine, M.M.; Ahtiainen, J.; Wågman, N.; Öberg, L.G.; Jørgensen, K.S. Fate and toxicity of chlorophenols, polychlorinated dibenzo-p-dioxins, and dibenzofurans during composting of contaminated sawmill soil. Environ. Sci. Technol. 1997, 31, 3244–3250. [Google Scholar] [CrossRef]
- Narihiro, T.; Kaiya, S.; Futamata, H.; Hiraishi, A. Removal of polychlorinated dioxins by semi-aerobic fed-batch composting with biostimulation of “Dehalococcoides”. J. Biosci. Bioeng. 2010, 109, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.N.; Stratton, G.W.; Murray, G. Effects of nutrient amendmentsand temperature on the biodegradationof pentachlorophenol contaminated soil. Water Air Soil Pollut. 2004, 151, 87–101. [Google Scholar] [CrossRef]
- Sinkkonen, A.; Kauppi, S.; Simpanen, S.; Rantalainen, A.L.; Strommer, R.; Romantschuk, M. Layer of organic pine forest soil on top of chlorophenol-contaminated mineral soil enhances contaminant degradation. Environ. Sci. Pollut. Res. 2013, 20, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.M.; Wu, M.J. Enhanced TCDD degradation by Fenton’s reagent preoxidation. J. Hazard. Mater. 2000, 74, 197–211. [Google Scholar] [CrossRef]
- Kao, C.M.; Chen, S.C.; Liu, J.K.; Wu, M.J. Evaluation of TCDD biodegradability under different redox conditions. Chemosphere 2001, 44, 1447–1454. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wu, J.H.; Lin, Y.Y.; Huang, H.J.; Chang, J.E. Bioremediation potential of soil contaminated with highly substituted polychlorinated dibenzo-p-dioxins and dibenzofurans: Microcosm study and microbial community analysis. J. Hazard. Mater. 2013, 261, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-Y.; Wu, J.-H.; Lin, S.-C.; Chang, J.-E. Bioremediation of polychlorinated-p-dioxins/dibenzofurans contaminated soil using simulated compost-amended landfill reactors under hypoxic conditions. J. Hazard. Mater. 2016, 312, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Binh, N.D.; Imsapsangworn, C.; Kim Oanh, N.T.; Parkpian, P.; Karstensen, K.; Giao, P.H.; DeLaune, R.D. Sequential anaerobic-aerobic biodegradation of 2,3,7,8-TCDD contaminated soil in the presence of CMC-coated nZVI and surfactant. Environ. Technol. 2016, 37, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Park, J.-W.; Häggblom, M.M. Enriching for microbial reductive dechlorination of polychlorinated dibenzo-p-dioxins and dibenzofurans. Environ. Pollut. 2014, 184, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Alkorta, I.; Garbisu, C. Phytoremediation of organic contaminants in soils. Bioresour. Technol. 2001, 79, 273–276. [Google Scholar] [CrossRef]
- Reichenauer, T.G.; Germida, J.J. Phytoremediation of organic contaminants in soil and groundwater. ChemSusChem 2008, 1, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Rezek, J.; in der Wiesche, C.; Macková, M.; Zadražil, F.; Macek, T. The effect of ryegrass (Lolium perenne) on decrease of PAH content in long term contaminated soil. Chemosphere 2008, 70, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Macek, T.; Macková, M.; Káš, J. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol. Adv. 2000, 18, 23–34. [Google Scholar] [CrossRef]
- Kurzawová, V.; Uhlík, O.; Macek, T.; Macková, M. Interactions of microbes and plants and their importance in phyto/rhizoremediation in PCB contaminated soil. Listy Cukrov. Reparske 2010, 126, 396–397. [Google Scholar]
- Wang, Y.; Oyaizu, H. Evaluation of the phytoremediation potential of four plant species for dibenzofuran-contaminated soil. J. Hazard. Mater. 2009, 168, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Oyaizu, H. Enhanced remediation of dioxins-spiked soil by a plant-microbe system using a dibenzofuran-degrading Comamonas sp. and Trifolium repens L. Chemosphere 2011, 85, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Futamata, H.; Uchida, T.; Yoshida, N.; Yonemitsu, Y.; Hiraishi, A. Distribution of dibenzofuran-degrading bacteria in soils polluted with different levels of polychlorinated dioxins. Microbes Environ. 2004, 19, 172–177. [Google Scholar] [CrossRef]
- Hanano, A.; Ammouneh, H.; Almousally, I.; Alorr, A.; Shaban, M.; Alnaser, A.A.; Ghanem, I. Traceability of polychlorinated dibenzo-dioxins/furans pollutants in soil and their ecotoxicological effects on genetics, functions and composition of bacterial community. Chemosphere 2014, 108, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Wu, J.H.; Chang, J.E. Pyrosequencing analysis reveals high population dynamics of the soil microcosm degrading octachlorodibenzofuran. Microbes Environ. 2014, 29, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Takahashi, N.; Hiraishi, A. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 2005, 71, 4325–4334. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A.; Kaiya, S.; Miyakoda, H.; Futamata, H. Biotransformation of polychlorinated dioxins and microbial community dynamics in sediment microcosms at different contamination levels. Microbes Environ. 2005, 20, 227–242. [Google Scholar] [CrossRef]
- Kaiya, S.; Utsunomiya, S.; Suzuki, S.; Yoshida, N.; Futamata, H.; Yamada, T.; Hiraishi, A. Isolation and functional gene analyses of aromatic-hydrocarbon-degrading bacteria from a polychlorinated-dioxin-dechlorinating process. Microbes Environ. 2012, 27, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Crowley, D.E.; Thompson, I.P. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 2003, 21, 123–130. [Google Scholar] [CrossRef]
- Chaudhry, Q.; Blom-Zandstra, M.; Gupta, S.; Joner, E.J. Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. Res. Int. 2005, 12, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Uhlík, O.; Musilová, L.; Rídl, J.; Hroudová, M.; Vlček, C.; Koubek, J.; Holečková, M.; Macková, M.; Macek, T. Plant secondary metabolite-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl. Microbiol. Biotechnol. 2013, 97, 9245–9256. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, P.K.; Hegde, R.S.; Fletcher, J.S. Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 1994, 28, 981–988. [Google Scholar] [CrossRef]
- Pham, T.T.; Pino Rodriguez, N.J.; Hijri, M.; Sylvestre, M. Optimizing polychlorinated biphenyl degradation by flavonoid-induced cells of the rhizobacterium Rhodococcus erythropolis U23A. PLoS ONE 2015, 10, e0126033. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, J.-P.; Pham, T.; Barriault, D.; Sylvestre, M. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl. Microbiol. Biotechnol. 2012, 95, 1589–1603. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Echartea, E.; Macek, T.; Demnerova, K.; Uhlik, O. Bacterial Biotransformation of Pentachlorophenol and Micropollutants Formed during Its Production Process. Int. J. Environ. Res. Public Health 2016, 13, 1146. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13111146
Lopez-Echartea E, Macek T, Demnerova K, Uhlik O. Bacterial Biotransformation of Pentachlorophenol and Micropollutants Formed during Its Production Process. International Journal of Environmental Research and Public Health. 2016; 13(11):1146. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13111146
Chicago/Turabian StyleLopez-Echartea, Eglantina, Tomas Macek, Katerina Demnerova, and Ondrej Uhlik. 2016. "Bacterial Biotransformation of Pentachlorophenol and Micropollutants Formed during Its Production Process" International Journal of Environmental Research and Public Health 13, no. 11: 1146. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13111146