Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam
Abstract
:1. Introduction
2. Materials and Methods
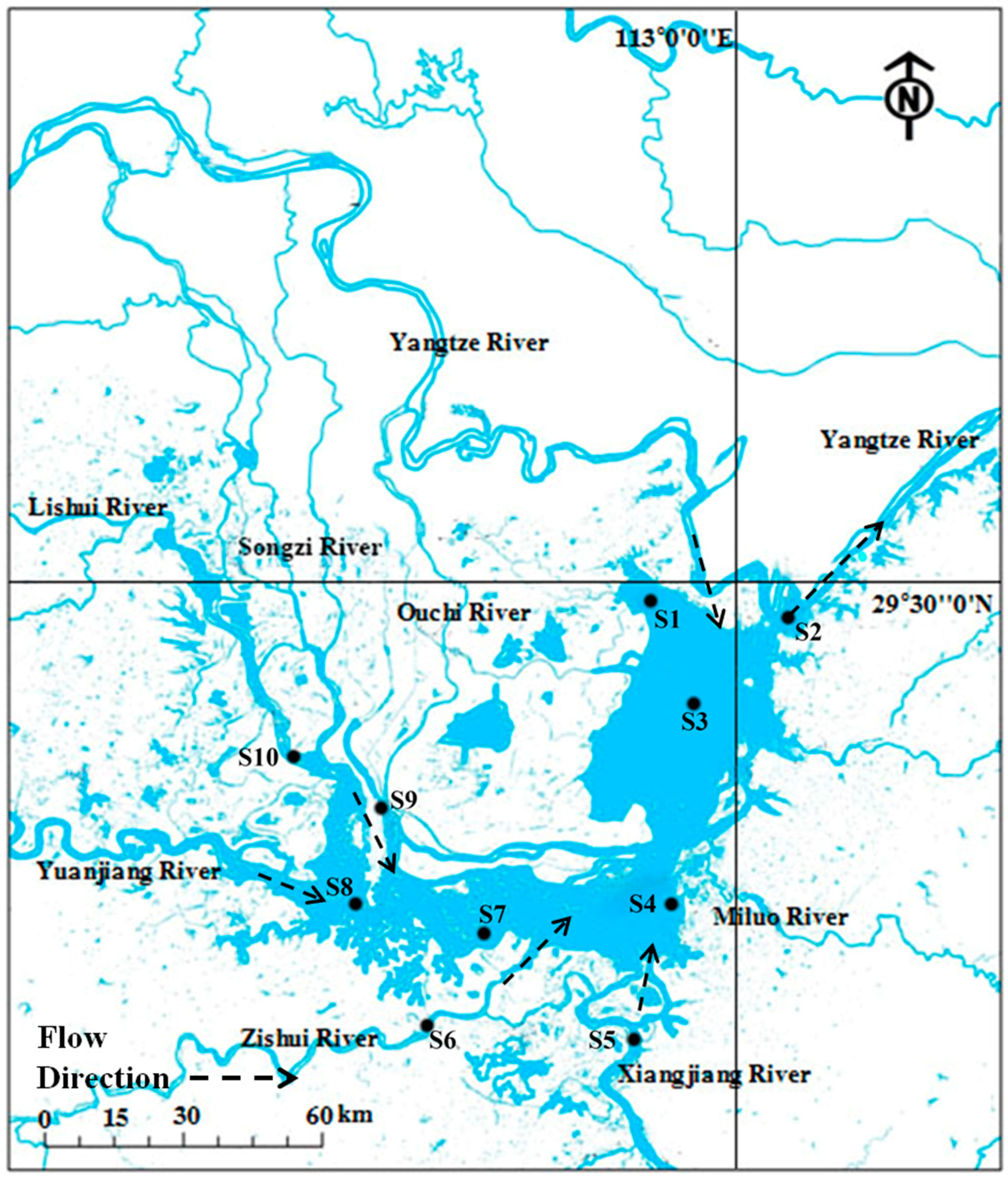
2.1. Site Description and Sediment Sampling
2.2. Analysis of Physicochemical Parameters
2.3. DNA Extraction
2.4. PCR Amplification of 16S rRNA Genes and Sequencing
2.5. Statistical Analysis
2.6. Accession Numbers
3. Results and Discussion
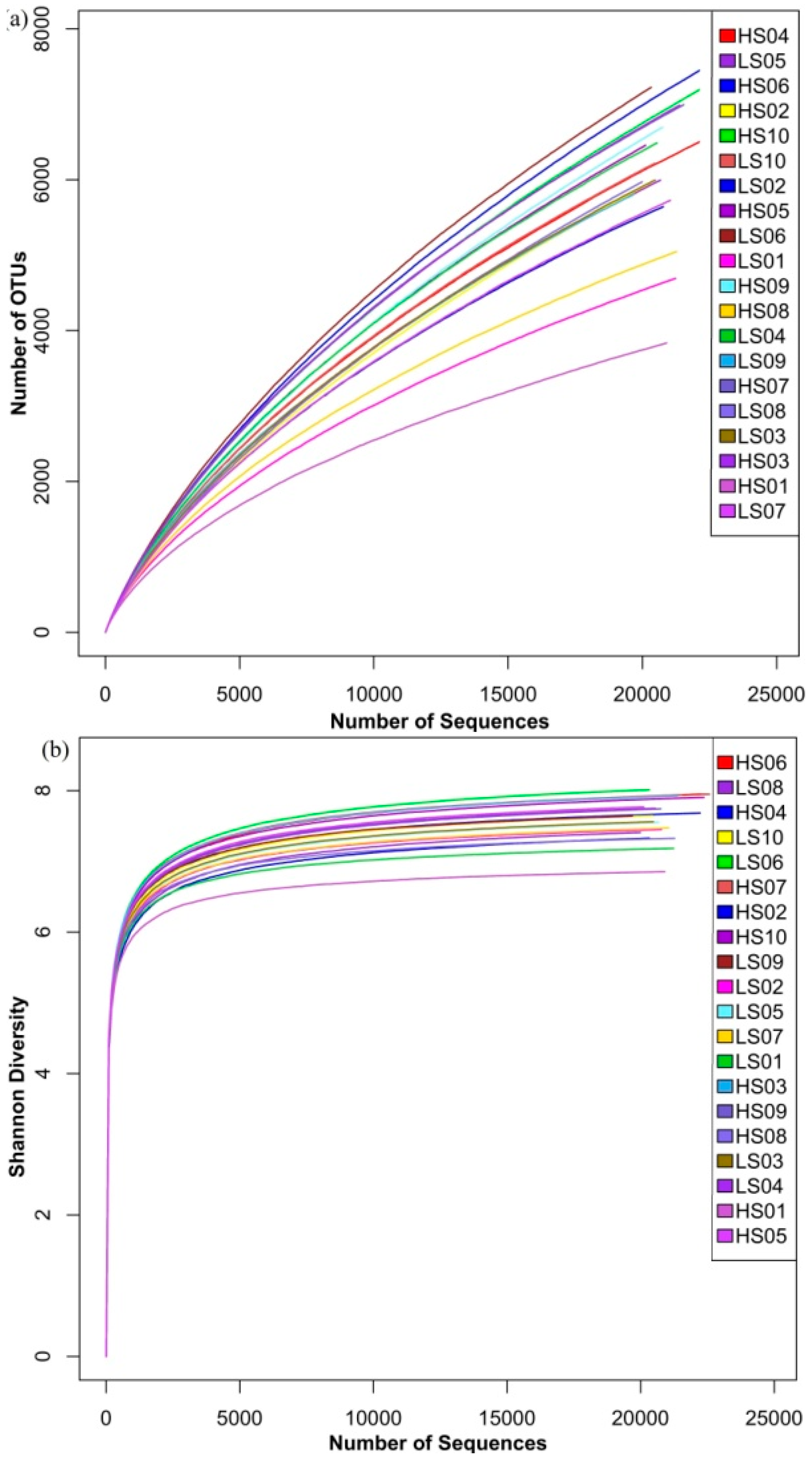
3.1. Richness and Diversity of Microbial Community
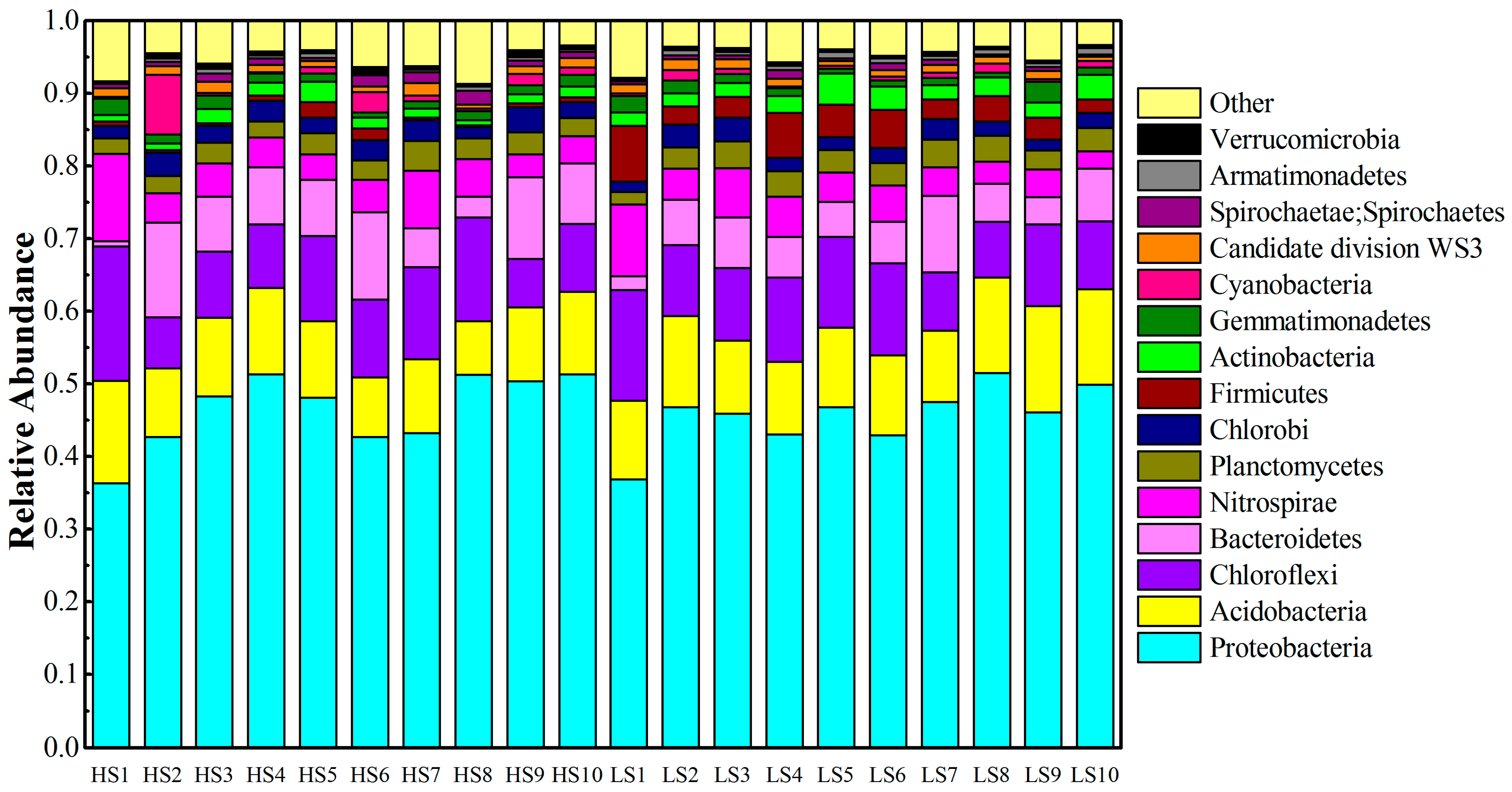
3.2. Phylum Level Taxonomic Distribution
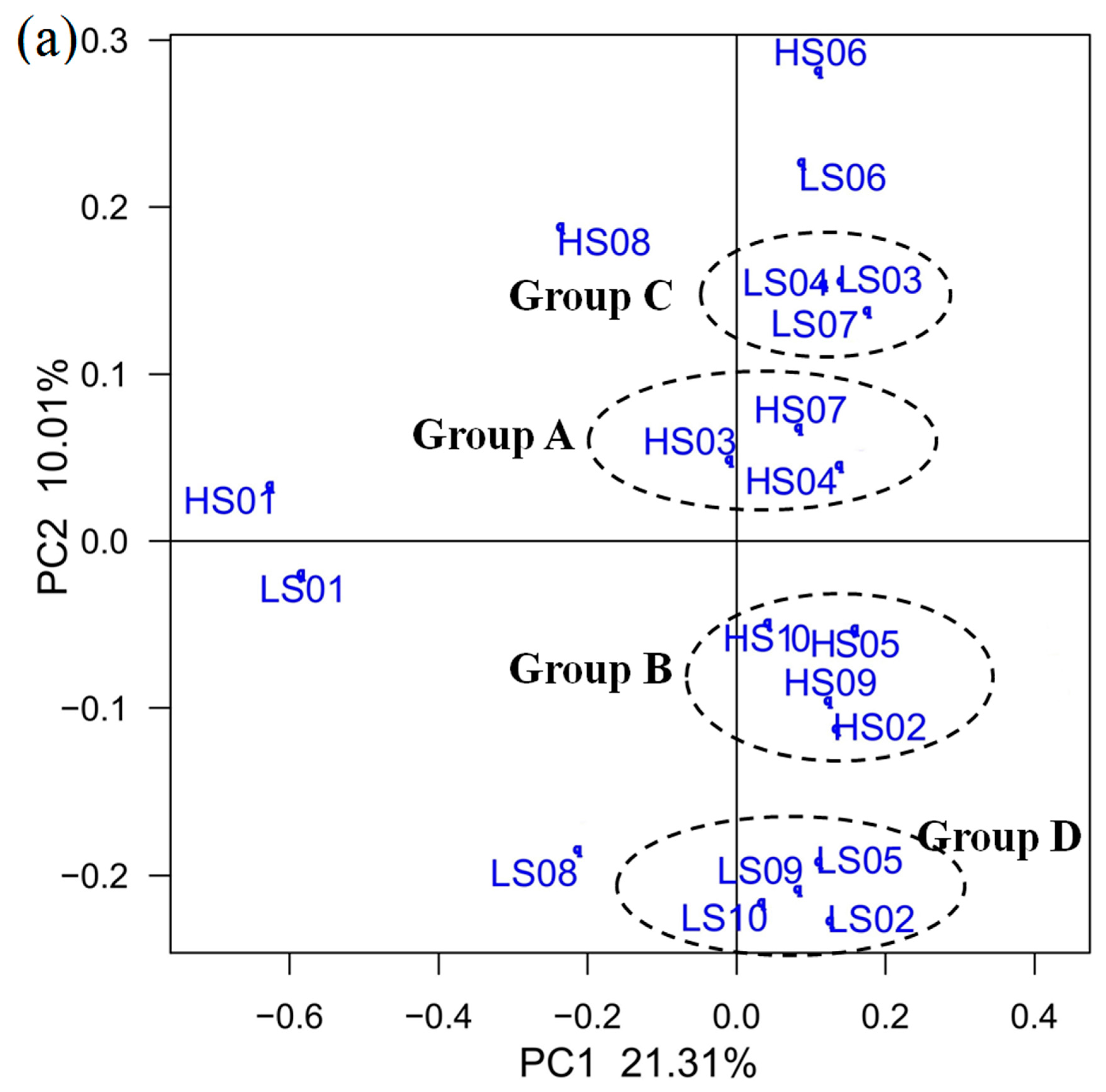
3.3. PCoA on OTU Level and RDA of Community Abundance on Dominant Bacterial Phyla
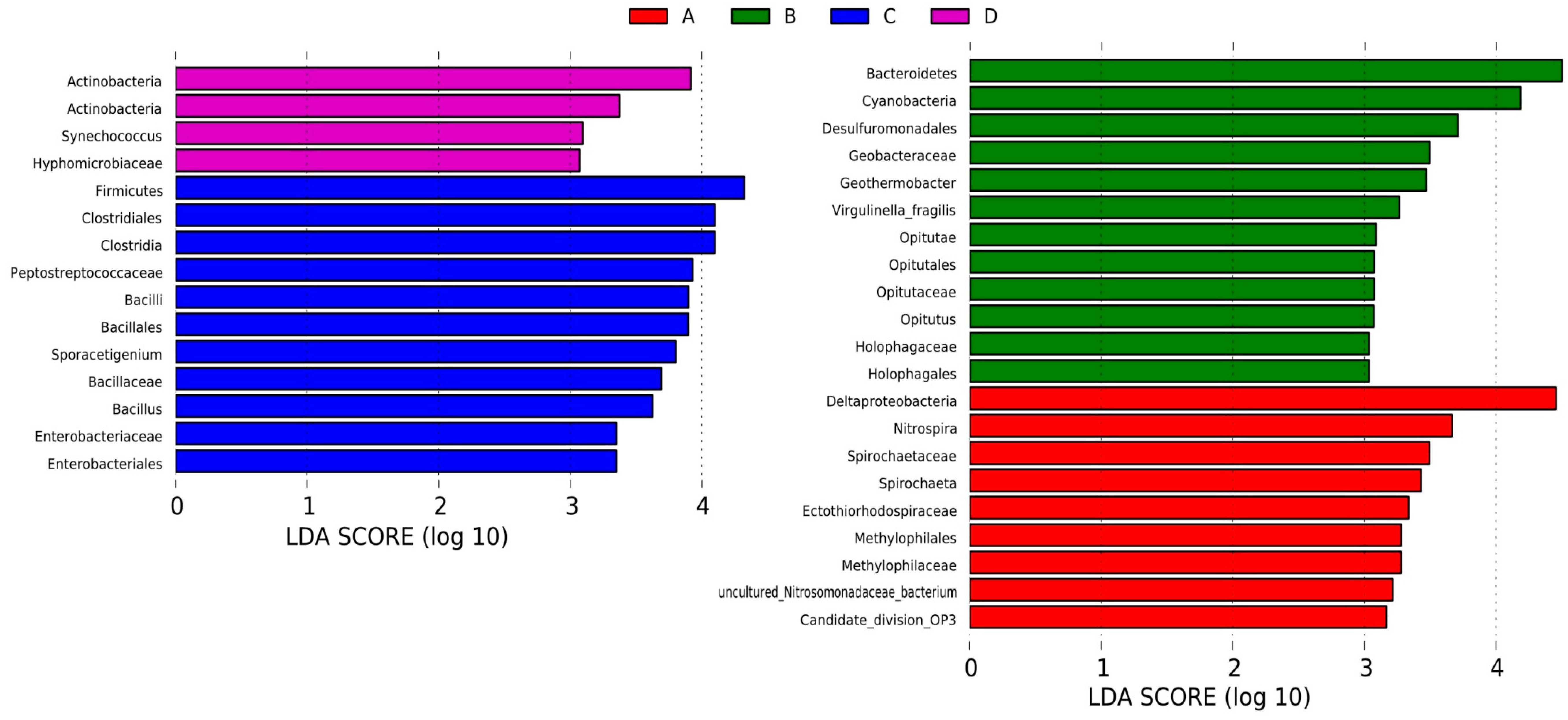
3.4. LEfSe Analysis Based on Community Abundance
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Walling, D.E. Human impact on land-ocean sediment transfer by the world’s rivers. Geomorphology 2006, 79, 192–216. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, J.; Zhang, Y.; Zhang, X.; Li, X. Enhanced lakebed sediment erosion in Dongting Lake induced by the operation of the Three Gorges Reservoir. J. Geog. Sci. 2015, 25, 917–929. [Google Scholar] [CrossRef]
- Chai, C.; Yu, Z.; Shen, Z.; Song, X.; Cao, X.; Yao, Y. Nutrient characteristics in the Yangtze River estuary and the adjacent East China Sea before and after impoundment of the Three Gorges Dam. Sci. Total Environ. 2009, 407, 4687–4695. [Google Scholar] [CrossRef] [PubMed]
- Bearsdley, R.C.; Limeburner, R.; Yu, H.; Cannon, G.A. Discharges of Changjiang (Yangtze River) into the East China Sea. Cont. Shelf Res. 1985, 4, 55–76. [Google Scholar]
- Nilsson, C.; Reidy, C.A.; Dynesius, M.; Revenga, C. Fragmentation and flowregulation of the world’s large river systems. Science 2005, 308, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.A.; Li, J.B.; Lu, D.Q.; Zhu, X.; Lu, C.Z.; Zhou, Y.Y.; Deng, C.X. The hydrological effect between Jingjiang River and Dongting Lake during the initial period of Three Gorges Project operation. J. Geog. Sci. 2010, 20, 771–786. [Google Scholar] [CrossRef]
- Zinger, L.; Amaral-Zettler, L.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Huse, S.M.; Welch, D.B.M.; Martiny, J.B.H.; Sogin, M.; Boetius, A.; Ramette, A. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE 2011, 6, e24570. [Google Scholar] [CrossRef] [PubMed]
- Nealson, K.H. Sediment bacteria: Who’s there, what are they doing, and what’s new? Annu. Rev. Earth Planet Sci. 1997, 25, 403–434. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.H.; Lin, G.H.; Gao, G.; Qin, B.Q.; Zhang, J.S.; Zhao, G.P.; Zhou, Z.H.; Shen, J.H. Bacterial and archaeal assemblages in sediments of a large shallow freshwater lake, Lake Taihu, as revealed by denaturing gradient gel electrophoresis. J. Appl. Microbiol. 2009, 106, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Dillon, J.G.; McMath, L.M.; Trout, A.L. Seasonal changes in bacterial diversity in the Salton Sea. Hydrobiologia 2009, 632, 49–64. [Google Scholar] [CrossRef]
- Chen, C.T.A. The Three Gorges Dam: Reducing the upwelling and thus productivity in the East China Sea. Geophys. Res. Lett. 2000, 27, 381–383. [Google Scholar] [CrossRef]
- Xu, K.H.; Milliman, J.D. Seasonal variations of sediment discharge from the Yangtze River before and after impoundment of the Three Gorges Dam. Geomorphology 2009, 104, 276–283. [Google Scholar] [CrossRef]
- Jiao, N.Z.; Zhang, Y.; Zeng, Y.H.; Gardner, W.D.; Mishonov, A.V.; Richardson, M.J.; Hong, N.; Pan, D.L.; Yan, X.H.; Jo, Y.H.; et al. Ecological anomalies in the East China Sea: Impacts of the Three Gorges Dam? Water Res. 2007, 41, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Bi, Y.; Deng, Y.; He, Z.L.; Wu, L.Y.; Van Nostrand, J.D.; Shi, Z.; Li, J.J.; Wang, X.; Hu, Z.Y.; et al. Impacts of the Three Gorges Dam on microbial structure and potential function. Sci. Rep. 2015, 5, 8605. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, H.; Watanabe, M.; Nakahara, T.; Xu, B.H.; Uchiyama, H. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 2002, 68, 5142–5150. [Google Scholar] [CrossRef]
- Yan, Q.Y.; Yu, Y.H.; Feng, W.S.; Yu, Z.G.; Chen, H.T. Plankton community composition in the Three Gorges Reservoir region revealed by PCR-DGGE and its relationships with environmental factors. J. Environ. Sci. 2008, 20, 732–738. [Google Scholar] [CrossRef]
- Gong, G.C.; Chang, J.; Chiang, K.P.; Hsiung, T.M.; Hung, C.C.; Duan, S.W.; Codispoti, L.A. Reduction of primary production and changing of nutrient ratio in the East China Sea: Effect of the Three Gorges Dam? Geophys. Res. Lett. 2006, 33, L07610. [Google Scholar] [CrossRef]
- Feng, L.; Hu, C.M.; Chen, X.L.; Zhao, X. Dramatic inundation changes of China’s two largest freshwater lakes linked to the Three Gorges Dam. Environ. Sci. Technol. 2013, 47, 9628–9634. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L. China scientists line up against dam that would alter protected wetlands. Science 2009, 326, 508–509. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zeng, G.; Liang, J.; Guo, S.; Dai, J.; Lu, L.; Wei, Z.; Xu, P.; Li, F.; Yuan, Y.; et al. Effect of early dry season induced by the Three Gorges Dam on the soil microbial biomass and bacterial community structure in the Dongting Lake wetland. Ecol. Indic. 2015, 53, 129–136. [Google Scholar] [CrossRef]
- Schutte, U.; Abdo, Z.; Bent, S.J.; Shyu, C.; Williams, C.J. Advances in the use of terminal restriction fragment length polymorphism (TRFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 2008, 80, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Cetecioglu, Z.; Ince, B.K.; Kolukirik, M.; Ince, O. Biogeographical distribution and diversity of bacterial and archaeal communities within highly polluted anoxic marine sediments from the Marmara sea. Mar. Pollut. Bull. 2009, 58, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Pratt, B.; Riesen, R.; Johnston, C.G. PLFA Analyses of microbial communities associated with PAH-contaminated riverbank sediment. Microb. Ecol. 2012, 64, 680–691. [Google Scholar] [CrossRef] [PubMed]
- Ruban, V.; Lopez-Sanchez, J.F.; Pardo, P.; Rauret, G.; Muntau, H.; Quevauviller, P. Development of a harmonised phosphorus extraction procedure and certification of a sediment reference material. J. Environ. Monit. 2001, 3, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Institute of Soil Science, Chinese Academy of Science. Soil Physics Chemical Analysis; Shanghai Science and Technology Press: Shanghai, China, 1978. [Google Scholar]
- Huang, W.; Lu, Y.; Li, J.H.; Zheng, Z.; Zhang, J.B.; Jiang, X. Effect of ionic strength on phosphorus sorption in different sediments from a eutrophic plateau lake. RSC Adv. 2015, 5, 79607–79615. [Google Scholar] [CrossRef]
- Liao, X.P.; Zhang, C.X.; Yao, L.L.; Li, J.L.; Liu, M.; Xu, L.; Evalde, M. Sorption behavior of nonylphenol (NP) on sewage-irrigated soil: kinetic and thermodynamic studies. Sci. Total Environ. 2014, 473, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Z.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008, 36, e120. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S Ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.B.; Ye, X.S.; Wang, K.; Chen, H.P.; Hu, C.J.; Zhu, J.L.; Zhang, D.M. Biogeography of the sediment bacterial community responds to a nitrogen pollution gradient in the East China Sea. Appl. Environ. Microbiol. 2014, 80, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Shi, Q.; Wen, D.; Li, Z.; Jefferson, W.A.; Feng, C.; Tang, X. Bacterial communities in the sediments of Dianchi Lake, a partitioned eutrophic waterbody in China. PLoS ONE 2012, 7, e37796. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, V.; Rehman, A.; Mishra, A.; Chauhan, P.S.; Nautiyal, C.S. Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 2012, 64, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, H.; Sekiguchi, Y.; Hanada, S.; Nakamura, K.; Nomura, N.; Matsumura, M.; Kamagata, Y. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl. Environ. Microbiol. 2005, 71, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.H.; Bai, Y.H.; Shi, Q.; Li, Z.X.; Sun, Q.H.; Sun, R.H.; Feng, C.P.; Tang, X.Y. Bacterial diversity in the polluted water of the Dianchi Lakeshore in China. Ann. Microbiol. 2012, 62, 715–723. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. R. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed]
- Gudasz, C.; Bastviken, D.; Premke, K.; Steger, K.; Tranvik, L.J. Constrained microbial processing of allochthonous organic carbon in boreal lake sediments. Limnol. Oceanogr. 2012, 57, 163–175. [Google Scholar] [CrossRef]


| Sample | TP (mg·kg−1) | TN (mg·kg−1) | Water Content (%) | OM (%) | pH |
|---|---|---|---|---|---|
| HS1 | 892.3 ± 10.2 | 620.2 ± 6.1 | 36.98 ± 2.13 | 8.21 ± 0.65 | 7.16 ± 0.44 |
| HS2 | 982.2 ± 9.2 | 1184.2 ± 8.2 | 44.55 ± 3.21 | 8.1 ± 0.49 | 7.35 ± 0.48 |
| HS3 | 722.4 ± 8.2 | 705.5 ± 8.1 | 47.31 ± 3.76 | 6.17 ± 0.46 | 7.25 ± 0.53 |
| HS4 | 823.2 ± 8.5 | 1004.6 ± 8.9 | 63.02 ± 4.29 | 7.62 ± 0.56 | 7.17 ± 0.49 |
| HS5 | 892.8 ± 8.6 | 992.1 ± 8.5 | 40.36 ± 5.75 | 5.97 ± 0.48 | 7.42 ± 0.61 |
| HS6 | 1003.7 ± 9.1 | 1197.8 ± 12.4 | 43.51 ± 3.98 | 7.07 ± 0.49 | 7.29 ± 0.67 |
| HS7 | 698.2 ± 7.3 | 821.2 ± 10.2 | 46.02 ± 3.57 | 5.16 ± 0.51 | 7.27 ± 0.62 |
| HS8 | 782.3 ± 7.8 | 995.3 ± 8.3 | 48.84 ± 3.43 | 7.03 ± 0.56 | 7.36 ± 0.68 |
| HS9 | 892.3 ± 7.7 | 1045.1 ± 10.3 | 35.11 ± 3.01 | 6.68 ± 0.41 | 7.46 ± 0.61 |
| HS10 | 602.3 ± 7.2 | 611.9 ± 8.3 | 30.76 ± 2.79 | 6.41 ± 0.62 | 7.37 ± 0.56 |
| LS1 | 921.2 ± 8.2 | 842.3 ± 7.3 | 33.21 ± 2.76 | 8.52 ± 0.69 | 7.21 ± 0.49 |
| LS2 | 1021.4 ± 9.2 | 1302.3 ± 8.1 | 40.12 ± 3.65 | 8.32 ± 0.72 | 7.43 ± 0.48 |
| LS3 | 823.1 ± 8.5 | 923.1 ± 9.4 | 44.21 ± 3.96 | 7.01 ± 0.67 | 7.31 ± 0.42 |
| LS4 | 937.5 ± 9.3 | 1198.5 ± 12.3 | 52.13 ± 4.02 | 7.34 ± 0.78 | 7.21 ± 0.51 |
| LS5 | 983.3 ± 9.1 | 1123.2 ± 12.8 | 36.21 ± 4.23 | 6.12 ± 0.51 | 7.39 ± 0.63 |
| LS6 | 1193.1 ± 10.2 | 1423.2 ± 11.3 | 38.97 ± 4.47 | 7.56 ± 0.68 | 7.33 ± 0.71 |
| LS7 | 821.5 ± 7.8 | 983.5 ± 11.7 | 40.46 ± 4.21 | 6.05 ± 0.59 | 7.35 ± 0.58 |
| LS8 | 842.1 ± 7.2 | 1045.2 ± 10.8 | 42.36 ± 3.76 | 7.12 ± 0.69 | 7.36 ± 0.62 |
| LS9 | 982.4 ± 8.7 | 1197.3 ± 11.5 | 30.21 ± 3.28 | 7.24 ± 0.66 | 7.41 ± 0.51 |
| LS10 | 701.2 ± 8.8 | 801.6 ± 10.3 | 27.98 ± 2.67 | 6.89 ± 0.57 | 7.51 ± 0.57 |
| Samples | Total Reads | OTU | Chao1 | ACE | Simpson | Shannon |
|---|---|---|---|---|---|---|
| HS1 | 20,903 | 3836 | 7863 | 10,727 | 0.0041 | 6.85 |
| LS1 | 21,231 | 4685 | 9569 | 13,925 | 0.0027 | 7.19 |
| HS2 | 20,323 | 5968 | 14,318 | 24,153 | 0.0056 | 7.33 |
| LS2 | 20,777 | 5631 | 11,931 | 17,402 | 0.0025 | 7.45 |
| HS3 | 21,389 | 6977 | 15,218 | 22,448 | 0.0012 | 7.92 |
| LS3 | 20,475 | 5964 | 13,828 | 21,097 | 0.0021 | 7.55 |
| HS4 | 22,239 | 6569 | 14,308 | 22,036 | 0.0018 | 7.75 |
| LS4 | 20,546 | 6511 | 14,138 | 21,108 | 0.0017 | 7.69 |
| HS5 | 20,118 | 6509 | 15,405 | 23,734 | 0.0016 | 7.77 |
| LS5 | 20,668 | 6048 | 13,522 | 20,455 | 0.0021 | 7.56 |
| HS6 | 22,586 | 7534 | 17,351 | 27,279 | 0.0014 | 8.12 |
| LS6 | 20,328 | 7193 | 16,320 | 25,817 | 0.0011 | 8.01 |
| HS7 | 21,547 | 7000 | 15,147 | 22,413 | 0.0012 | 7.93 |
| LS7 | 21,042 | 5709 | 12,116 | 18,138 | 0.0021 | 7.47 |
| HS8 | 21,271 | 5047 | 14,636 | 23,396 | 0.0022 | 7.48 |
| LS8 | 19,989 | 5963 | 9953 | 14,672 | 0.0031 | 7.32 |
| HS9 | 20,751 | 6670 | 16,138 | 25,942 | 0.0017 | 7.74 |
| LS9 | 19,691 | 5750 | 12,903 | 20,436 | 0.0016 | 7.63 |
| HS10 | 22,367 | 7233 | 16,542 | 25,599 | 0.0013 | 7.90 |
| LS10 | 20,452 | 6159 | 13,562 | 20,807 | 0.0020 | 7.64 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Jiang, X. Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam. Int. J. Environ. Res. Public Health 2016, 13, 617. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13060617
Huang W, Jiang X. Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam. International Journal of Environmental Research and Public Health. 2016; 13(6):617. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13060617
Chicago/Turabian StyleHuang, Wei, and Xia Jiang. 2016. "Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam" International Journal of Environmental Research and Public Health 13, no. 6: 617. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph13060617






