Diversity and Antimicrobial Resistance Genotypes in Non-Typhoidal Salmonella Isolates from Poultry Farms in Uganda
Abstract
:1. Introduction
2. Materials and Methods
2.1. The NTS isolate collection
2.2. Pulsed-Field Gel Electrophoresis (PFGE) and Bionumerics Analysis
2.3. Bacterial DNA Extraction
2.4. Detection of Integrons and Antibiotic Resistance Genes
3. Results
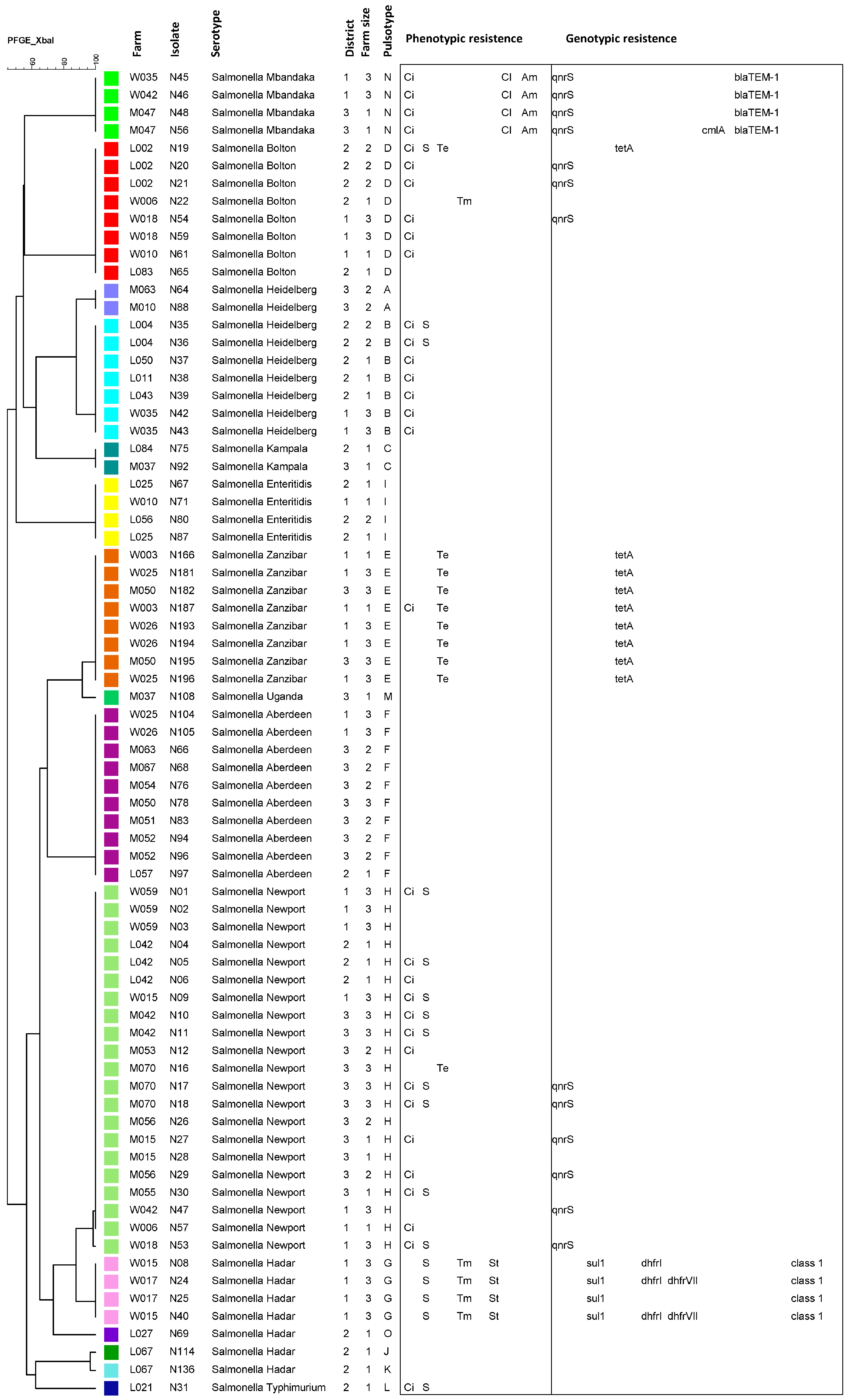
3.1. Pulsed-Field Gel Electrophoresis Typing
3.2. Detection of Integrons and Antibiotic Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Camps, N.; Dominguez, A.; Company, M.; Perez, M.; Pardos, J.; Llobet, T.; Usera, M.A.; Salleras, L.; Working Group for the Investigation of the Outbreak of Salmonellosis. A foodborne outbreak of Salmonella infection due to overproduction of egg-containing foods for a festival. Epidemiol. Infect. 2005, 133, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Shi, Z.; Wei, J.; Ma, Z. A brief review of foodborne zoonoses in China. Epidemiol. Infect. 2011, 139, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Dallal, M.M.S.; Taremi, M.; Gachkar, L.; Modarressi, S.; Sanaei, M.; Bakhtiari, R.; Yazdi, M.K.S.; Zali, M.R. Characterization of antibiotic resistant patterns of Salmonella serotypes isolated from beef and chicken samples in Tehran. Jundishapur J. Microbiol. 2009, 2, 124–131. [Google Scholar]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Mourao, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. International Collaboration on Enteric Disease ‘Burden of Illness, S.; The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg. Infect. Dis. 2015, 21, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.M.; Molyneux, E.M.; Walsh, A.L.; Cheesbrough, J.S.; Molyneux, M.E.; Hart, C.A. Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr. Infect. Dis. J. 2000, 19, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A.; Graham, S.M.; Walsh, A.L.; Wilson, L.; Phiri, A.; Molyneux, E.; Zijlstra, E.E.; Heyderman, R.S.; Hart, C.A.; Molyneux, M.E. Epidemics of invasive Salmonella enterica serovar enteritidis and S-enterica serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 2008, 46, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, G.M.; Curtis, N. Non-typhoidal Salmonella in Children: Microbiology, Epidemiology and Treatment. In Hot Topics in Infection and Immunity in Children IX; Curtis, N., Finn, A., Pollard, A.J., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Kariuki, S.; Gordon, M.A.; Feasey, N.; Parry, C.M. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 2015, 33, S21–S29. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, S.; Revathi, G.; Kariuki, N.; Kiiru, J.; Mwituria, J.; Hart, C.A. Characterisation of community acquired non-typhoidal Salmonella from bacteraemia and diarrhoeal infections in children admitted to hospital in Nairobi, Kenya. BMC Microbiol. 2006, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Gaffga, N.H.; Behravesh, C.B.; Ettestad, P.J.; Smelser, C.B.; Rhorer, A.R.; Cronquist, A.B.; Comstock, N.A.; Bidol, S.A.; Patel, N.J.; Gerner-Smidt, P.; et al. Outbreak of Salmonellosis Linked to Live Poultry from a Mail-Order Hatchery. N. Engl. J. Med. 2012, 366, 2065–2073. [Google Scholar] [CrossRef] [PubMed]
- Lynne, A.M.; Rhodes-Clark, B.S.; Bliven, K.; Zhao, S.H.; Foley, S.L. Antimicrobial resistance genes associated with Salmonella enterica serovar Newport isolates from food animals. Antimicrob. Agents Chemother. 2008, 52, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Huang, J.H.; Wu, Q.P.; Zhang, J.M.; Liu, S.R.; Guo, W.P.; Cai, S.Z.; Yu, S.B. Prevalence, antimicrobial resistance and genetic diversity of Salmonella isolated from retail ready-to-eat foods in China. Food Control 2016, 60, 50–56. [Google Scholar] [CrossRef]
- Tamang, M.D.; Nam, H.M.; Kim, T.S.; Jang, G.C.; Jung, S.C.; Lim, S.K. Emergence of Extended-Spectrum beta-Lactamase (CTX-M-15 and CTX-M-14)-Producing Nontyphoid Salmonella with Reduced Susceptibility to Ciprofloxacin among Food Animals and Humans in Korea. J. Clin. Microbiol. 2011, 49, 2671–2675. [Google Scholar] [CrossRef] [PubMed]
- Chuanchuen, R.; Padungtod, P. Antimicrobial Resistance Genes in Salmonella enterica Isolates from Poultry and Swine in Thailand. J. Vet. Med. Sci. 2009, 71, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Lunguya, O.; Lejon, V.; Phoba, M.F.; Bertrand, S.; Vanhoof, R.; Glupczynski, Y.; Verhaegen, J.; Muyembe-Tamfum, J.J.; Jacobs, J. Antimicrobial Resistance in Invasive Non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-spectrum Beta Lactamases. PLoS Negl. Trop. Dis. 2013, 7, e2103. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Alshalchi, S.; Breimhurst, P.; Munoz-Aguayo, J.; Flores-Figueroa, C.; Vidovic, S. Strong influence of livestock environments on the emergence and dissemination of distinct multidrug-resistant phenotypes among the population of non-typhoidal Salmonella. PLoS ONE 2017, 12, e0179005. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Van Nhiem, D.; Paulsen, P.; Suriyasathaporn, W.; Smulders, F.J.M.; Kyule, M.N.; Baumann, M.P.O.; Zessin, K.H.; Ngan, P.H. Preliminary analysis of tetracycline residues in marketed pork in Hanoi, Vietnam. In Impact of Emerging Zoonotic Diseases on Animal Health; Blouin, E.F., Maillard, J.C., Eds.; New York Academy of Sciences: New York, NY, USA, 2006. [Google Scholar]
- Abdel-Maksoud, M.; Abdel-Khalek, R.; El-Gendy, A.; Gamal, R.F.; Abdelhady, H.M.; House, B.L. Genetic characterisation of multidrug-resistant Salmonella enterica serotypes isolated from poultry in Cairo, Egypt. Afr. J. Lab. Med. 2015, 4. [Google Scholar] [CrossRef]
- Tabo, D.A.; Diguimbaye, C.D.; Granier, S.A.; Moury, F.; Brisabois, A.; Elgroud, R.; Millemann, Y. Prevalence and antimicrobial resistance of non-typhoidal Salmonella serotypes isolated from laying hens and broiler chicken farms in N’Djamena, Chad. Vet. Microbiol. 2013, 166, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Characterization of integrons and resistance genes in multidrug-resistant Salmonella enterica isolated from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014, 189, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Fashae, K.; Ogunsola, F.; Aarestrup, F.M.; Hendriksen, R.S. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J. Infect. Dev. Ctries. 2010, 4, 484–494. [Google Scholar] [PubMed]
- Dione, M.M.; Ieven, M.; Garin, B.; Marcotty, T.; Geerts, S. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Broiler Farms, Chicken Carcasses, and Street-Vended Restaurants in Casamance, Senegal. J. Food Prot. 2009, 72, 2423–2427. [Google Scholar] [CrossRef] [PubMed]
- Elgroud, R.; Zerdoumi, F.; Benazzouz, M.; Bouzitouna-Bentchouala, C.; Granier, S.A.; Fremy, S.; Brisabois, A.; Dufour, B.; Millemann, Y. Characteristics of Salmonella Contamination of Broilers and Slaughterhouses in the Region of Constantine (Algeria). Zoonoses Public Health 2009, 56, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, R.; Abbassi, M.S.; Garcia, V.; Garcia-Fierro, R.; Fernandez, J.; Kilani, H.; Jaouani, I.; Khayeche, M.; Messadi, L.; Rodicio, M.R. Antimicrobial drug resistance and genetic properties of Salmonella enterica serotype Enteritidis circulating in chicken farms in Tunisia. J. Infect. Public Health 2017, 10, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Odoch, T.; Wasteson, Y.; L’Abée-Lund, T.; Muwonge, A.; Kankya, C.; Nyakarahuka, L.; Tegule, S.; Skjerve, E. Prevalence, antimicrobial susceptibility and risk factors associated with non-typhoidal Salmonella on Ugandan layer hen farms. BMC Vet. Res. 2017, 13, 365. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization (ISO). ISO 6579:2002/Amd 1:2007 Annex D: Detection of Salmonella spp. in Animal Faeces and in Environmental Samples from the Primary Production; ISO: Geneva, Switzerland, 2007. [Google Scholar]
- Ahmed, A.M.; Hussein, A.I.A.; Shimamoto, T. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 2007, 59, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Motoi, Y.; Sato, M.; Maruyama, M.; Watanabe, H.; Fukumoto, Y.; Shimamoto, T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007, 73, 6686–6690. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Butaye, P.; Cloeckaert, A.; Schwarz, S. Genes and mutations conferring antimicrobial resistance in Salmonella: An update. Microbes Infect. 2006, 8, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Bacci, C.; Boni, E.; Alpigiani, I.; Lanzoni, E.; Bonardi, S.; Brindani, F. Phenotypic and genotypic features of antibiotic resistance in Salmonella enterica isolated from chicken meat and chicken and quail carcasses. Int. J. Food Microbiol. 2012, 160, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Lariviere, S.; Harel, J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149 : K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, B.W.; Wu, Y.; Zhang, Z.F.; Meng, X.F.; Xi, M.L.; Wang, X.; Xia, X.D.; Shi, X.M.; Wang, D.P.; et al. Molecular characterization of Salmonella enterica serovar Enteritidis on retail raw poultry in six provinces and two National cities in China. Food Microbiol. 2015, 46, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Bin Kim, H.; Park, C.H.; Kim, C.J.; Kim, E.C.; Jacoby, G.A.; Hooper, D.C. Prevalence of Plasmid-Mediated Quinolone Resistance Determinants over a 9-Year Period. Antimicrob. Agents Chemother. 2009, 53, 639–645. [Google Scholar]
- Randall, L.P.; Cooles, S.W.; Osborn, M.K.; Piddock, L.J.V.; Woodward, M.J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004, 53, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, S.H.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.C.; McDermott, P.F.; Ayers, S.; Meng, J.H. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Poppe, C.; Martin, L.; Muckle, A.; Archambault, M.; McEwen, S.; Weir, E. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can. J. Vet. Res. 2006, 70, 105–114. [Google Scholar] [PubMed]
- Afema, J.A.; Byarugaba, D.K.; Shah, D.H.; Atukwase, E.; Nambi, M.; Sischo, W.M. Potential Sources and Transmission of Salmonella and Antimicrobial Resistance in Kampala, Uganda. PLoS ONE 2016, 11, e0152130. [Google Scholar] [CrossRef] [PubMed]
- Andoh, L.A.; Dalsgaard, A.; Obiri-Danso, K.; Newman, M.J.; Barco, L.; Olsen, J.E. Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol. Infect. 2016, 144, 3288–3299. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, E.; Gros-Claude, J.D.P.; Rivoal, K.; Rose, V.; Tall, F.; Mead, G.C.; Salvat, G. Epidemiological analysis of Salmonella enterica ssp enterica serovars Hadar, Brancaster and Enteritidis from humans and broiler chickens in Senegal using pulsed-field gel electrophoresis and antibiotic susceptibility. J. Appl. Microbiol. 2005, 99, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Thomason, B.M.; Biddle, J.W.; Cherry, W.B. Detection of salmonellae in environment. Appl. Microbiol. 1975, 30, 764–767. [Google Scholar] [PubMed]
- Winfield, M.D.; Groisman, E.A. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 3687–3694. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica Serotypes and Food Commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Powers, J.H.; Chiller, T.M.; Aidara-Kane, A.; Aarestrup, F.M. World Health Organization Ranking of Antimicrobials According to Their Importance in Human Medicine: A Critical Step for Developing Risk Management Strategies for the Use of Antimicrobials in Food Production Animals. Clin. Infect. Dis. 2009, 49, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Nakano, H.; Shimamoto, T. Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: Description of an unusual class 2 integron. J. Antimicrob. Chemother. 2005, 55, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Agerso, Y.; Aarestrup, F.M.; dos Reis, E.M.F.; Rodrigues, D.D. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. J. Antimicrob. Chemother. 2006, 58, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Guerri, M.L.; Aladuena, A.; Echeita, A.; Rotger, R. Detection of integrons and antibiotic-resistance genes in Salmonella enterica serovar Typhimurium, isolates with resistance to ampicillin and variable susceptibility to amoxicillin-clavulanate. Int. J. Antimicrob. Agents 2004, 24, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Ata, Z.; Yibar, A.; Arslan, E.; Mustak, K.; Gunaydin, E. Plasmid-mediated quinolone resistance in Salmonella serotypes isolated from chicken carcasses in Turkey. Acta Vet BRNO 2014, 83, 281–286. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kwon, Y.K.; Tamang, M.D.; Jang, H.K.; Jeong, O.M.; Lee, H.S.; Kang, M.S. Plasmid-Mediated Quinolone Resistance in Escherichia coli Isolates from Wild Birds and Chickens in South Korea. Microb. Drug Resist. 2016, 22, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Geetha, V.K.; Yugendran, T.; Srinivasan, R.; Harish, B.N. Plasmid-mediated quinolone resistance in typhoidal Salmonellae: A preliminary report from South India. Indian J. Med. Microbiol. 2014, 32, 31–34. [Google Scholar] [PubMed]
- Hopkins, K.L.; Davies, R.H.; Threlfall, E.J. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int. J. Antimicrob. Agents 2005, 25, 358–373. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.V.; Gole, V.C.; McWhorter, A.R.; Abraham, S.; Chousalkar, K.K. Antimicrobial resistance of non-typhoidal Salmonella isolates from egg layer flocks and egg shells. Int. J. Food Microbiol. 2015, 203, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Miko, A.; Pries, K.; Schroeter, A.; Helmuth, R. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 2005, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Murgia, M.; Bouchrif, B.; Timinouni, M.; Al-Qahtani, A.; Al-Ahdal, M.N.; Cappuccinelli, P.; Rubino, S.; Paglietti, B. Antibiotic resistance determinants and genetic analysis of Salmonella enterica isolated from food in Morocco. Int. J. Food Microbiol. 2015, 215, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.A.; El-Hofy, F.I.; Shafik, S.M.; Abdelrahman, M.A.; Elsaid, G.A. Characterization of Virulence-Associated Genes, Antimicrobial Resistance Genes, and Class 1 Integrons in Salmonella enterica serovar Typhimurium Isolates from Chicken Meat and Humans in Egypt. Foodborne Pathog. Dis. 2016, 13, 281–288. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, H.; Tahoun, A.; El-Gohary, A.; El-Abasy, M.; El-Khayat, F.; Gillespie, T.; Kitade, Y.; Hafez, H.M.; Neubauer, H.; El-Adawy, H. Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on acquired tetracycline resistance genes. Fems Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE). Annual Report on the Use of Antimicrobial Agents in Animals-Better Understanding of the Global Situation; OIE: Paris, France, 2015. [Google Scholar]
- Abatcha, M.G.; Zakaria, Z.; Gurmeet, K.D.; Thong, K.T. Antibiograms, Resistance Genes, Class I Integrons and PFGE profiles of Zoonotic Salmonella in Malaysia. Trop. Biomed. 2015, 32, 573–586. [Google Scholar]
- Berendsen, B.; Stolker, L.; de Jong, J.; Nielen, M.; Tserendorj, E.; Sodnomdarjaa, R.; Cannavan, A.; Elliott, C. Evidence of natural occurrence of the banned antibiotic chloramphenicol in herbs and grass. Anal. Bioanal. Chem. 2010, 397, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Zhang, Q.; Alali, W.Q.; Wang, J.W.; Meng, L.Y.; Xiao, Y.P.; Yang, H.; Chen, S.; Cui, S.H.; Yang, B.W. Characterization of extended-spectrum beta-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 2017, 248, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Eguale, T.; Birungi, J.; Asrat, D.; Njahira, M.N.; Njuguna, J.; Gebreyes, W.A.; Gunn, J.S.; Djikeng, A.; Engidawork, E. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob. Resist. Infect. Control 2017, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Giuriatti, J.; Stefani, L.M.; Brisola, M.C.; Crecencio, R.B.; Bitner, D.S.; Faria, G.A. Salmonella Heidelberg: Genetic profile of its antimicrobial resistance related to extended spectrum beta-lactamases (ESBLs). Microb. Pathog. 2017, 109, 195–199. [Google Scholar] [CrossRef] [PubMed]
| Target Category | Target Gene | Primer Sequence | Amplicon Size (bp) | Annealing Temp (°C) | Reference |
|---|---|---|---|---|---|
| Integron | Class 1 integron | Variable size | 55 | [33] | |
| 5’-CS | GGCATCCAAGCAGCAAG | ||||
| 3’-CS | AAGCAGACTTGACCTGA | ||||
| Class 2 integron | 491 | 55 | [33] | ||
| hep74 | CGGGATCCCGGACGGCATGCACGATTTGTA | ||||
| hep51 | GATGCCATCGCAAGTACGAG | ||||
| Resistance to ampicillin by detection of four β-lactamase genes | blaPSE-1 | CGCTTCCCGTTAACAAGTAC | 419 | 57 | [35] |
| CTGGTTCATTTCAGATAGCG | |||||
| blaCMY-2 | TGGCCAGAACTGACAGGCAAA | 462 | 64 | [36] | |
| TTTCTCCTGAACGTGGCTGGC | |||||
| blaTEM-1 | AGGAAGAGTATGATTCAACA | 535 | 55 | [37] | |
| CTCGTCGTTTGGTATGGC | |||||
| blaOxA | ACCAGATTCAACTTTCAA | 590 | 55 | [38] | |
| TCTTGGCTTTTATGCTTG | |||||
| Resistance to ciprofloxacin by detection of four fluoroquinolone plasmid mediated quinolone resistance genes | qnrA | AGAGGATTTCTCACGCCAGG | 580 | 54 | [39] |
| TGCCAGGCACAGATCTTGAC | |||||
| qnrB | GATCGTGAAAGCCAGAAAGG | 476 | 53 | [40] | |
| ATGAGCAACGATGCCTGGTA | |||||
| qnrC | GGGTTGTACATTTATTGAATCG | 307 | 53 | [40] | |
| CACCTACCCATTTATTTTCA | |||||
| qnrS | GCAAGTTCATTGAACAGGGT | 428 | 54 | [39] | |
| TCTAAACCGTCGAGTTCGGCG | |||||
| Resistance to chloramphenicol by detection of four phenicol resistance genes | floR | AACCCGCCCTCTGGATCAAGTCAA | 548 | 60 | [41] |
| CAAATCACGGGCCACGCTGTATC | |||||
| cat1 | CTTGTCGCCTTGCGTATAAT | 508 | 55 | [42] | |
| ATCCCAATGGCATCGTAAAG | |||||
| cat2 | AACGGCATGATGAACCTGAA | 547 | 55 | [42] | |
| ATCCCAATGGCATCGTAAAG | |||||
| cmlA | CGCCACGGTGTTGTTGTTAT | 394 | 55 | [42] | |
| GCGACCTGCGTAAATGTCAC | |||||
| Resistance to sulfonamide by detection of two dihydropteroate reductase genes | sul1 | GCG CGG CGT GGG CTA CCT | 350 | 65 | [43] |
| GATTTCCGCGACACCGAGACAA | |||||
| sul2 | CGG CAT CGT CAA CAT AACC | 720 | 52 | [43] | |
| GTG TGC GGA TGA AGT CAG | |||||
| Resistance to tetracycline by detection of three efflux pump genes | tetA | GCTACATCCTGCTTGCCTTC | 210 | 55 | [35] |
| CATAGATCGCCGTGAAGAGG | |||||
| tetB | TTGGTTAGGGGCAAGTTTTG | 659 | 55 | [35] | |
| GTAATGGGCCAATAACACCG | |||||
| tetG | CAG CTTTCG GATTCT TACGG | 844 | 55 | [35] | |
| GAT TGGTGA GGCTCG TTAGC | |||||
| Resistance to trimethoprim by detection of five dihydrofolate reductase genes | dhfrI | AAGAATGGAGTTATCGGGAATG | 391 | 50 | [37] |
| GGGTAAAAACTGGCCTAAAATTG | |||||
| dhfrV | CTGCAAAAGCGAAAAACGG | 432 | 50 | [37] | |
| AGCAATAGTTAATGTTTGAGCTAAAG | |||||
| dhfrVII | GGTAATGGCCCTGATATCCC | 265 | 50 | [37] | |
| TGTAGATTTGACCGCCACC | |||||
| dhfrIX | TCTAAACATGATTGTCGCTGTC | 452 | 50 | [37] | |
| TTGTTTTCAGTAATGGTCGGG | |||||
| dhfrXIII | CAGGTGAGCAGAAGATTTTT | 294 | 50 | [37] | |
| CCTCAAAGGTTTGATGTACC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odoch, T.; Sekse, C.; L’Abee-Lund, T.M.; Høgberg Hansen, H.C.; Kankya, C.; Wasteson, Y. Diversity and Antimicrobial Resistance Genotypes in Non-Typhoidal Salmonella Isolates from Poultry Farms in Uganda. Int. J. Environ. Res. Public Health 2018, 15, 324. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15020324
Odoch T, Sekse C, L’Abee-Lund TM, Høgberg Hansen HC, Kankya C, Wasteson Y. Diversity and Antimicrobial Resistance Genotypes in Non-Typhoidal Salmonella Isolates from Poultry Farms in Uganda. International Journal of Environmental Research and Public Health. 2018; 15(2):324. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15020324
Chicago/Turabian StyleOdoch, Terence, Camilla Sekse, Trine M. L’Abee-Lund, Helge Christoffer Høgberg Hansen, Clovice Kankya, and Yngvild Wasteson. 2018. "Diversity and Antimicrobial Resistance Genotypes in Non-Typhoidal Salmonella Isolates from Poultry Farms in Uganda" International Journal of Environmental Research and Public Health 15, no. 2: 324. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15020324




