Continuous Flow-Constructed Wetlands for the Treatment of Swine Waste Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Sampling
2.2. Enumeration E. coli from MPN Method
2.3. Isolation of Potential Pathogenic E. coli from Wetland
2.4. Typing of E. coli Using REP-PCR
2.5. Analysis of E. coli Genotyping
2.6. Statistical Analysis
3. Results
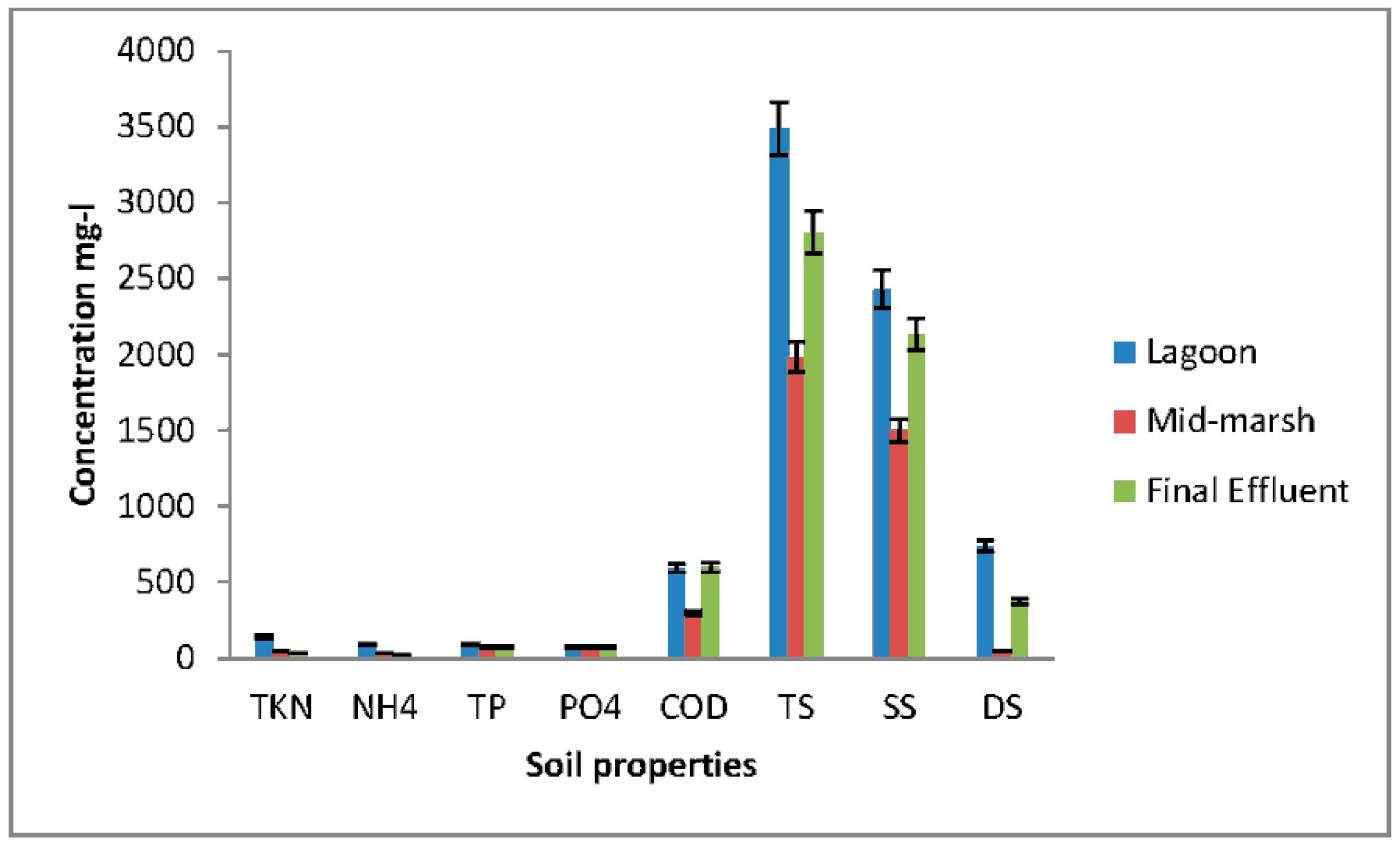
3.1. Removal E. coli Isolates and Nutrients from Wetland Samples
3.2. Characterization of Potentially Pathogenic E. coli
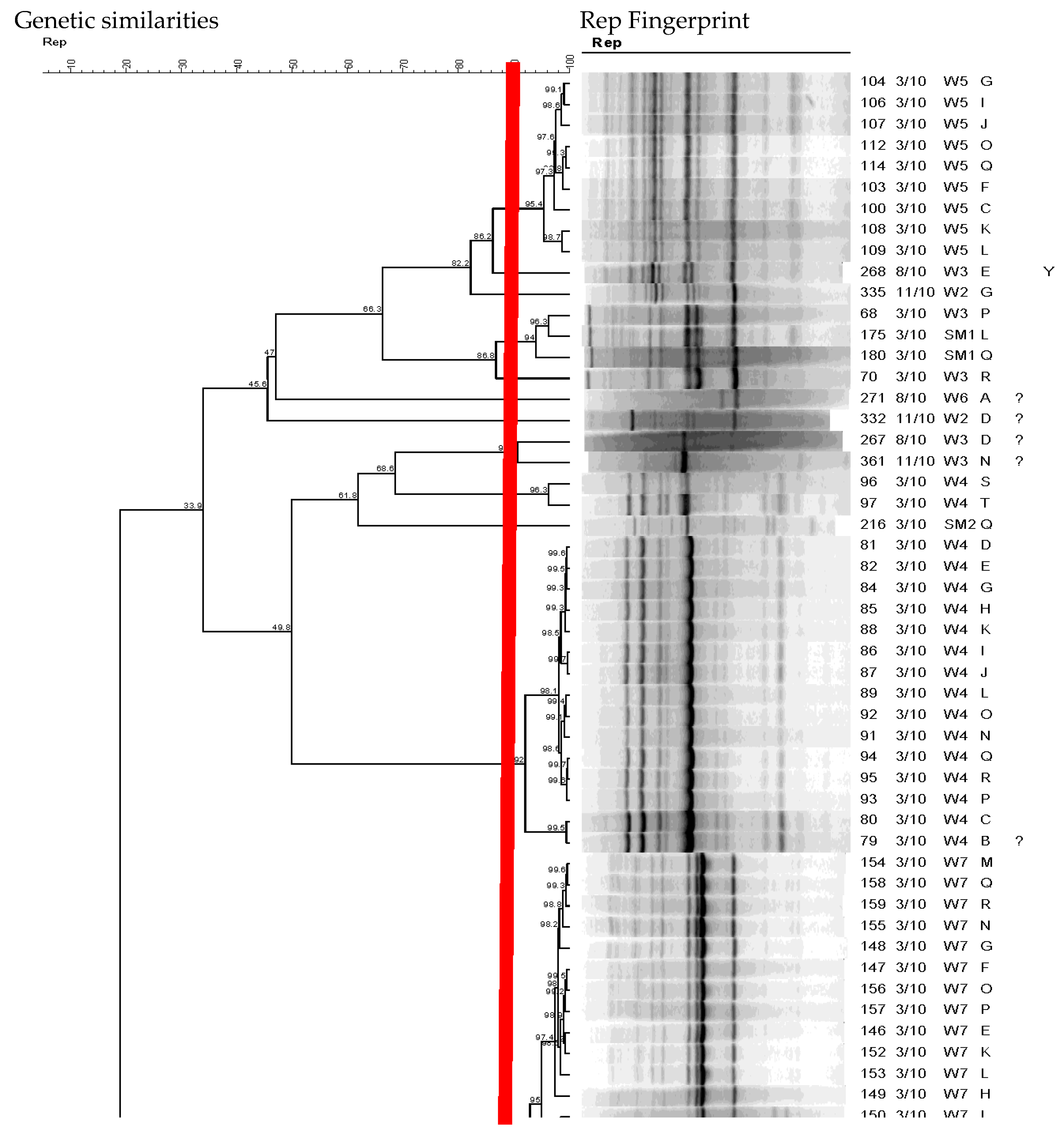
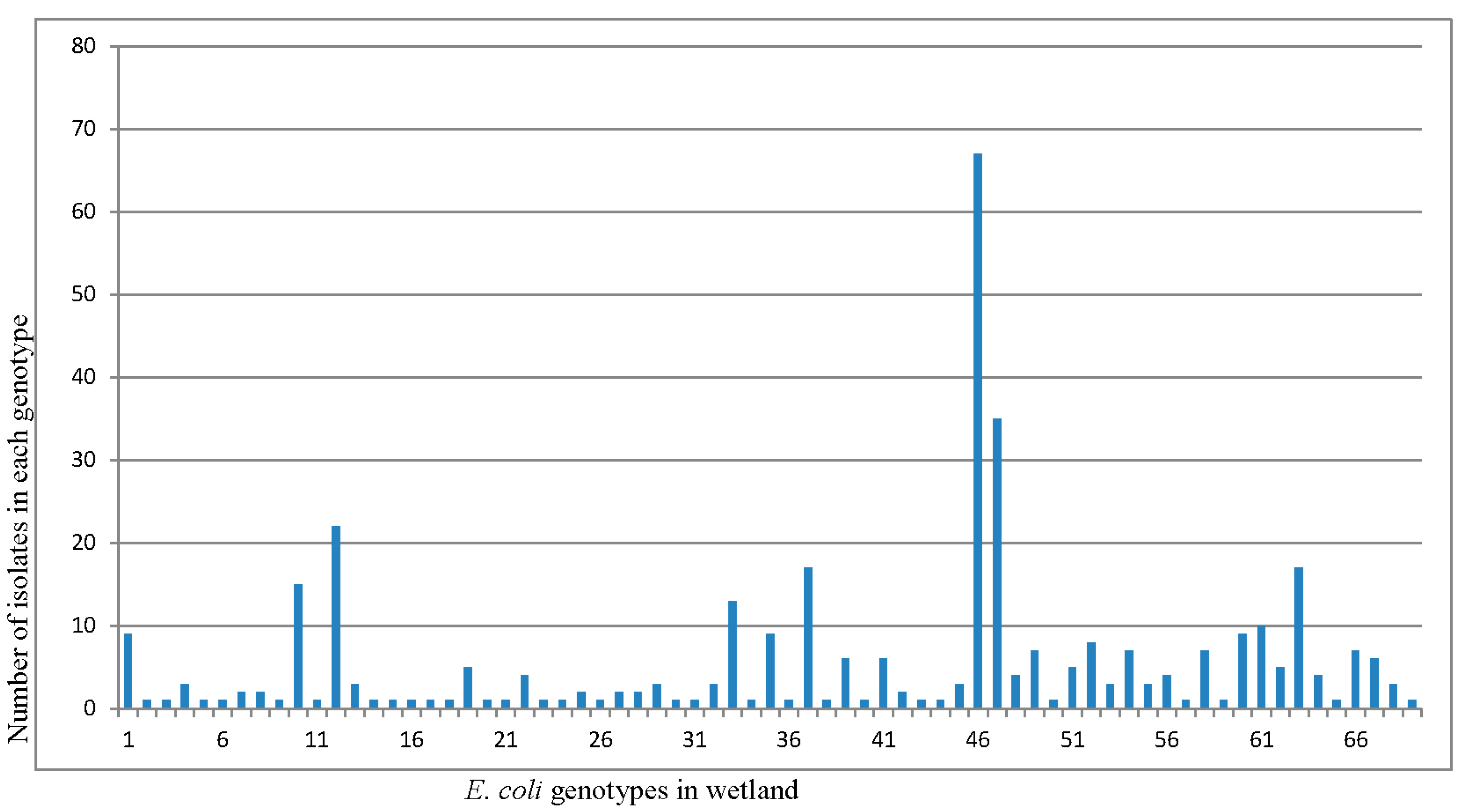
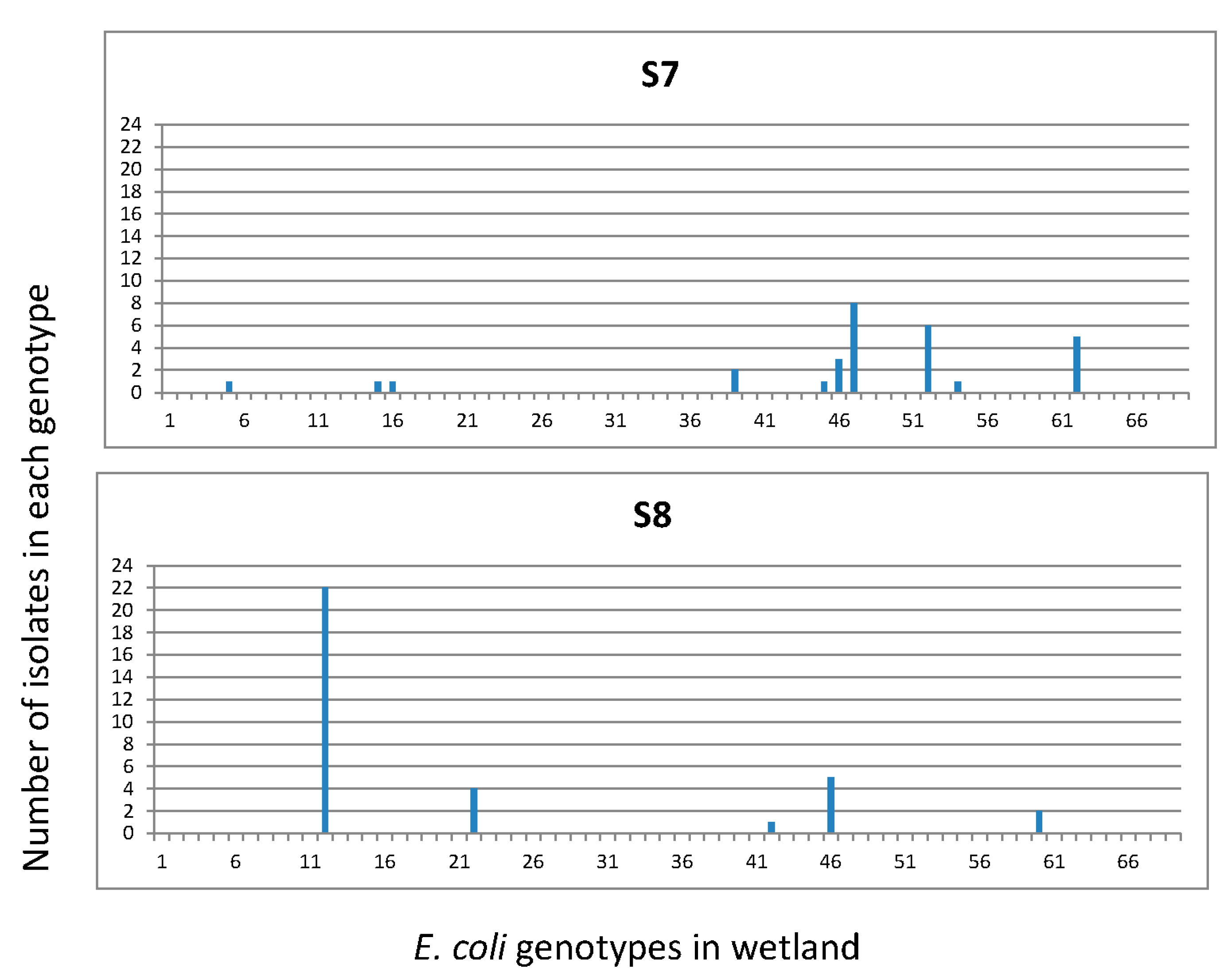
3.3. Diversity of E. coli Isolates in the Continuous Flow-Constructed Wetland with REP-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kadlec, R.H.; Knight, R.L. Treatment Wetlands; Lewis Publishers: New York, NY, USA, 1996. [Google Scholar]
- Ibekwe, A.M.; Grieve, C.M.; Lyon, S.R. Characterization of microbial communities and composition in constructed dairy wetland wastewater effluent. Appl. Environ. Microbiol. 2003, 69, 5060–5069. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Lyon, S.R.; Leddy, M.; Jacobson-Meyer, M. Impact of plant density and microbial composition on water quality from a free water surface constructed wetlands. J. Appl. Microbiol. 2007, 102, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Casteel, M.J.; Sobsey, M.D.; Mueller, J.P. Fecal contamination of agricultural soils before and after hurricane-associated flooding in North Carolina. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2006, 41, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.B.; Hunt, R.G.; Phillips, R.; Stone, K.; Grubbs, A. Treatment of swine wastewater in marsh-pond-marsh constructed wetlands. Water Sci. Technol. 2001, 44, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Reddy, G.B. Soil bacterial communities in constructed wetlands treated with swine wastewater using PCR-DGGE technique. Bioresour. Technol. 2010, 101, 11541–11549. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Sadowsky, M.J. Escherichia coli in the environment: Implication for water quality and human health. Microbes Environ. 2008, 23, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Ma, J.; Murinda, S.; Reddy, G.B. Microbial diversity in continuous flow constructed wetland for the treatment of swine waste. Hydrol. Curr. Res. 2017, 8, 277. [Google Scholar] [CrossRef]
- Eaton, A.D.; Clesceri, L.S.; Greenberg, A.E.; Franson, M.A.H. Standard Method for Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Paton, J.C.; Paton, A.W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 1998, 11, 450–479. [Google Scholar] [PubMed]
- DebRoy, C.; Maddox, C.W. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev 2001, 1, 129–140. [Google Scholar] [CrossRef]
- Dombek, P.E.; Johnson, L.K.; Zimmerley, S.T.; Sadowsky, M.J. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 2000, 66, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Yan, T. Genotypic diversity of Escherichia coli in the water and soil of tropical watersheds in Hawaii. Appl. Environ. Microbiol. 2011, 77, 3988–3997. [Google Scholar] [CrossRef] [PubMed]
- Lyautey, E.; Lu, Z.; Lapen, D.R.; Wilkes, G.; Scott, A.; Berkers, T.; Edge, T.A.; Topp, E. Distribution and diversity of Escherichia coli populations in the South Nation river drainage basin, eastern Ontario, Canada. Appl. Environ. Microbiol. 2010, 76, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, J.L.W.; de Bruijn, F.J. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis. In DNA Markers: Protocols, Applications, and Overviews; Caetano-Anollés, G., Gresshoff, P.M., Eds.; Wiley and Sons: New York, NY, USA, 1997; pp. 151–171. [Google Scholar]
- Stern, M.J.; Ames, G.F.L.; Smith, N.H.; Robinson, E.C.; Higgins, C.F. Repetitive extragenic palindromic sequences: A major component of the bacterial genome. Cell 1984, 37, 1015–1026. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS/STAT User’s Guide, Release 9.1; SAS Institute Inc.: Cary, NC, USA, 2009. [Google Scholar]
- Chandran, A.; Mazumder, A. Pathogenic potential, genetic diversity, and population structure of Escherichia coli strains isolated from a forest-dominated watershed (Comox Lake) in British Columbia, Canada. Appl. Environ. Microbiol. 2015, 81, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Poach, M.E.; Hunt, P.G.; Reddy, G.B.; Stone, K.C.; Johnson, M.H.; Grubbs, A. Swine wastewater treatment by marsh-pond-marsh constructed wetlands under varying nitrogen loads. Ecol. Eng. 2004, 23, 165–175. [Google Scholar] [CrossRef]
- Chapman, T.A.; Wu, X.Y.; Barchia, I.; Bettelheim, K.A.; Driesen, S.; Trott, D.; Wilson, M.; Chin, J.J. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 2006, 72, 4782–4795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frigon, D.; Biswal, B.K.; Masson, A.; Gehr, R. Biological and physicochemical wastewater treatment processes reduce the prevalence of virulent Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Murinda, S.E.; DebRoy, C.; Reddy, G.B. Potential pathogens, antimicrobial patterns and genotypic diversity of Escherichia coli isolates in constructed wetlands treating swine wastewater. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Graves, A.K.; Weaver, R.W. Characterization of enterococci populations collected from a subsurface flow constructed wetland. J. Appl. Microbiol. 2010, 108, 1226–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steer, D.; Fraser, L.; Boddy, J.; Seibert, B. Efficiency of small constructed wetlands for subsurface treatment of single-family domestic effluent. Ecol. Eng. 2002, 18, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Vymazal, J.; Kropfelova, L. Wastewater Treatment in Constructed Wetlands with Horizontal Sub-Surface Flow; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar]
- Sidrach-Cardona, R.; Bécares, E. Fecal indicator bacteria resistance to antibiotics in experimental constructed wetlands. Ecol. Eng. 2013, 50, 107–111. [Google Scholar] [CrossRef]
- Call, D.R.; Borucki, M.K.; Loge, F.J. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods 2003, 53, 235–243. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Kruse, H.; Tast, E.; Hammerum, A.M.; Jensen, L.B. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 2000, 6, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Whitman, R.L.; Przybyla-Kelly, K.; Shively, A.D.; Nevers, M.B.; Byappanahalli, M.N. Sunlight, season, snowmelt, storm, and source affect E. coli populations in an artificially ponded stream. Sci. Total Environ. 2008, 390, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Duriez, P.; Topp, E. Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial farm. Appl. Environ. Microbiol. 2007, 73, 5486–5493. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B.; Diez-Gonzalez, F.; Jarvis, G.N. Effects of diet shifts on Escherichia coli in cattle. J. Dairy Sci. 2000, 83, 863–873. [Google Scholar] [CrossRef]
- McLellan, S.L. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 2004, 70, 4658–4665. [Google Scholar] [CrossRef] [PubMed]



| Month | Sample Name | O Type | H Type | LT | Sta | STb | STX1 | STX2 | EAE | CNF1 | CNF2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| March | S1C | - | 43 | - | - | - | - | - | - | - | - |
| March | S1F | - | 43 | - | - | - | - | - | - | - | - |
| March | S1G | - | 43 | - | - | - | - | - | - | - | - |
| March | S1U | - | 19 | - | + | + | - | - | - | - | - |
| March | S1W | - | 4 | - | + | + | - | - | - | - | - |
| March | S2C | - | 11 | - | - | - | - | - | + | - | - |
| March | S2D | - | 11 | - | - | - | - | - | + | - | - |
| March | S2E | - | 11 | - | - | - | - | - | + | - | - |
| March | S2K | - | 43 | - | - | - | - | - | - | - | - |
| March | S2P | 88 | 38 | - | - | - | - | - | - | - | - |
| March | S2Y | - | 4 | - | + | + | - | - | - | - | - |
| March | S2Z | - | 4 | - | + | + | - | - | - | - | - |
| March | S2AA | - | 4 | - | + | + | - | - | - | - | - |
| March | S2AB | - | 4 | - | + | + | - | - | - | - | - |
| March | S2AC | - | 4 | - | + | + | - | - | - | - | - |
| March | S3W | - | 4 | - | + | + | - | - | - | - | - |
| March | S3X | - | 9 | - | + | - | - | - | - | - | - |
| March | S3Y | - | 4 | - | + | + | - | - | - | - | - |
| March | SM1A | - | 36 | - | + | + | - | + | - | - | - |
| March | SM1C | - | 36 | - | + | + | - | + | - | - | - |
| March | SM1T | - | 36 | - | + | + | - | + | - | - | - |
| March | SM1U | - | 36 | - | + | + | - | + | - | - | - |
| March | SM1V | - | 36 | - | + | + | - | + | - | - | - |
| March | SM2N | 98 | 5 | - | - | + | - | - | - | - | - |
| March | SM2U | - | 36 | - | + | + | - | + | - | - | - |
| March | SM2V | - | 36 | - | + | + | - | + | - | - | - |
| March | SM2W | - | 11 | - | - | - | - | - | + | - | - |
| March | SM2X | - | 36 | - | + | + | - | + | - | - | - |
| March | SM2Y | - | 11 | - | - | - | - | - | + | - | - |
| March | SM2Z | - | 19 | - | + | + | - | - | - | - | - |
| March | PW1A | - | 19 | - | + | + | - | - | - | - | - |
| March | PW1B | - | 19 | - | + | + | - | - | - | - | - |
| March | PW1C | - | 19 | - | + | + | - | - | - | - | - |
| March | PW1D | - | 19 | - | + | + | - | - | - | - | - |
| March | PW2A | - | 19 | - | + | + | - | - | - | - | - |
| March | PW2B | - | 19 | - | + | + | - | - | - | - | - |
| March | PW2C | - | 19 | - | + | + | - | - | - | - | - |
| March | PW2D | - | 19 | - | + | + | - | - | - | - | - |
| March | PW2E | - | 19 | - | + | + | - | - | - | - | - |
| March | PW3A | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2E | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2F | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2G | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2H | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2I | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2J | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2K | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2L | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2M | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2N | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2O | - | 19 | - | + | + | - | - | - | - | - |
| March | PSM2P | - | 19 | - | + | + | - | - | - | - | - |
| August | S1C | - | 11 | - | - | - | - | - | + | - | - |
| August | S1H | - | 11 | - | - | - | - | - | + | - | - |
| August | S1J | - | 11 | - | - | - | - | - | + | - | - |
| August | S1K | - | 11 | - | - | - | - | - | + | - | - |
| August | S2B | - | 32 | - | - | - | - | - | - | - | - |
| August | S3E | 178 | + | - | - | - | - | - | - | - | - |
| November | S1L | - | 11 | - | - | - | - | - | + | - | - |
| November | S2K | - | 11 | - | - | - | - | - | + | - | - |
| November | S2L | - | 11 | - | - | - | - | - | + | - | - |
| November | S3A | - | + | - | - | - | - | - | - | - | - |
| November | S3B | 2 | + | - | - | - | - | - | - | - | - |
| November | S3C | - | 4 | - | - | - | - | - | - | - | - |
| November | S3G | - | 11 | - | - | - | - | - | + | - | - |
| November | S3H | - | 11 | - | - | - | - | - | + | - | - |
| November | S3J | - | 11 | - | - | - | - | - | + | - | - |
| November | S3O | - | 4 | + | + | - | - | - | - | - | - |
| November | S6K | - | 11 | - | - | - | - | - | + | - | - |
| November | SM1G | - | 30 | - | + | - | - | + | - | - | - |
| November | SM1H | - | 30 | - | + | - | - | + | - | - | - |
| November | SM1L | - | 30 | - | + | - | - | + | - | - | - |
| Season | No. of Isolates | No. of Unique Genotypes | H′ Index | Frequency Ratio |
|---|---|---|---|---|
| March | 247 | 40 | 3.173 | 0.16 |
| August | 61 | 18 | 2.335 | 0.28 |
| November | 113 | 34 | 3.019 | 0.30 |
| Total | 421 | 92 | 3.231 | 0.25 |
| Sampling Locations | No. of Isolates | No. of Unique Genotypes | H′ Index | Frequency Ratio |
|---|---|---|---|---|
| swine house effluent (S1) | 135 | 31 | 2.976 | 0.23 |
| primary lagoon 1 (S2) | 57 | 18 | 2.569 | 0.31 |
| secondary lagoon 2 (S3) | 57 | 17 | 2.177 | 0.30 |
| storage tank (S4) | 48 | 20 | 2.779 | 0.42 |
| continuous wetland influent (S5) | 24 | 7 | 1.310 | 0.29 |
| continuous wetland effluent (S6) | 30 | 6 | 1.705 | 0.20 |
| storage pond (S7) | 36 | 10 | 2.090 | 0.27 |
| land application (S8) | 34 | 5 | 1.086 | 0.15 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibekwe, A.M.; Murinda, S.E. Continuous Flow-Constructed Wetlands for the Treatment of Swine Waste Water. Int. J. Environ. Res. Public Health 2018, 15, 1369. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15071369
Ibekwe AM, Murinda SE. Continuous Flow-Constructed Wetlands for the Treatment of Swine Waste Water. International Journal of Environmental Research and Public Health. 2018; 15(7):1369. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15071369
Chicago/Turabian StyleIbekwe, Abasiofiok M., and Shelton E. Murinda. 2018. "Continuous Flow-Constructed Wetlands for the Treatment of Swine Waste Water" International Journal of Environmental Research and Public Health 15, no. 7: 1369. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph15071369




