Mechanism of Membrane Fouling Control by HMBR: Effect of Microbial Community on EPS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Raw Wastewater Characteristics
2.3. Operational Conditions
2.4. Analytical Methods
2.5. Microbial Community Analysis
- (1)
- DNA extraction. The DNA was extracted from the samples using the MoBio PowerSoil DNA extraction kit (MO BIO Laboratories, Carlsbad, CA, USA) following the manufacturer’s instructions.
- (2)
- Polymerase chain reaction (PCR) amplification. PCR amplification of 16S rRNA genes was performed using general bacterial primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGTCAATTCMTTTGAGTTT-3′). The primers also contained the Illumina 5′overhang adapter sequences for two-step amplicon library building.
- (3)
- Miseq HTS. The barcoded PCR products were purified using a DNA gel extraction kit (Axygen, China) and quantified using the FTC-3000 TM real-time PCR. The libraries were sequenced by 2 × 300 bp paired-end sequencing on the MiSeq platform using MiSeq v3 Reagent Kit (Illumina, San Diego, CA, USA) at TinyGene Bio-Tech (Shanghai) Co., Ltd., China.
- (4)
- Bioinformatic analysis. The raw fastq files were demultiplexed based on the barcode. Paired-end (PE) reads for all samples were run through Trimmomatic (version 0.35) to remove low-quality base pairs. Trimmed reads were then merged using FLASH program (version 1.2.11). The low quality contigs were removed based on screen.seqs command in mothur (version 1.33.3). The cleaned reads were clustered at 97% sequence identity into operational taxonomic units (OTUs) using the UPARSE pipeline (usearch version v8.1.1756). The OTU representative sequences were assigned for taxonomy against Silva 128 database by the classify.seqs command in mothur. Taxonomies (from phylum to species) of the OTUs were determined depending on National Center for Biotechnology Information. Based on the taxonomy, the statistical analysis of community structure was carried out at the level of phylum, class, order, family, genus and species.
3. Results and Discussions
3.1. Overall Set-Up Performance
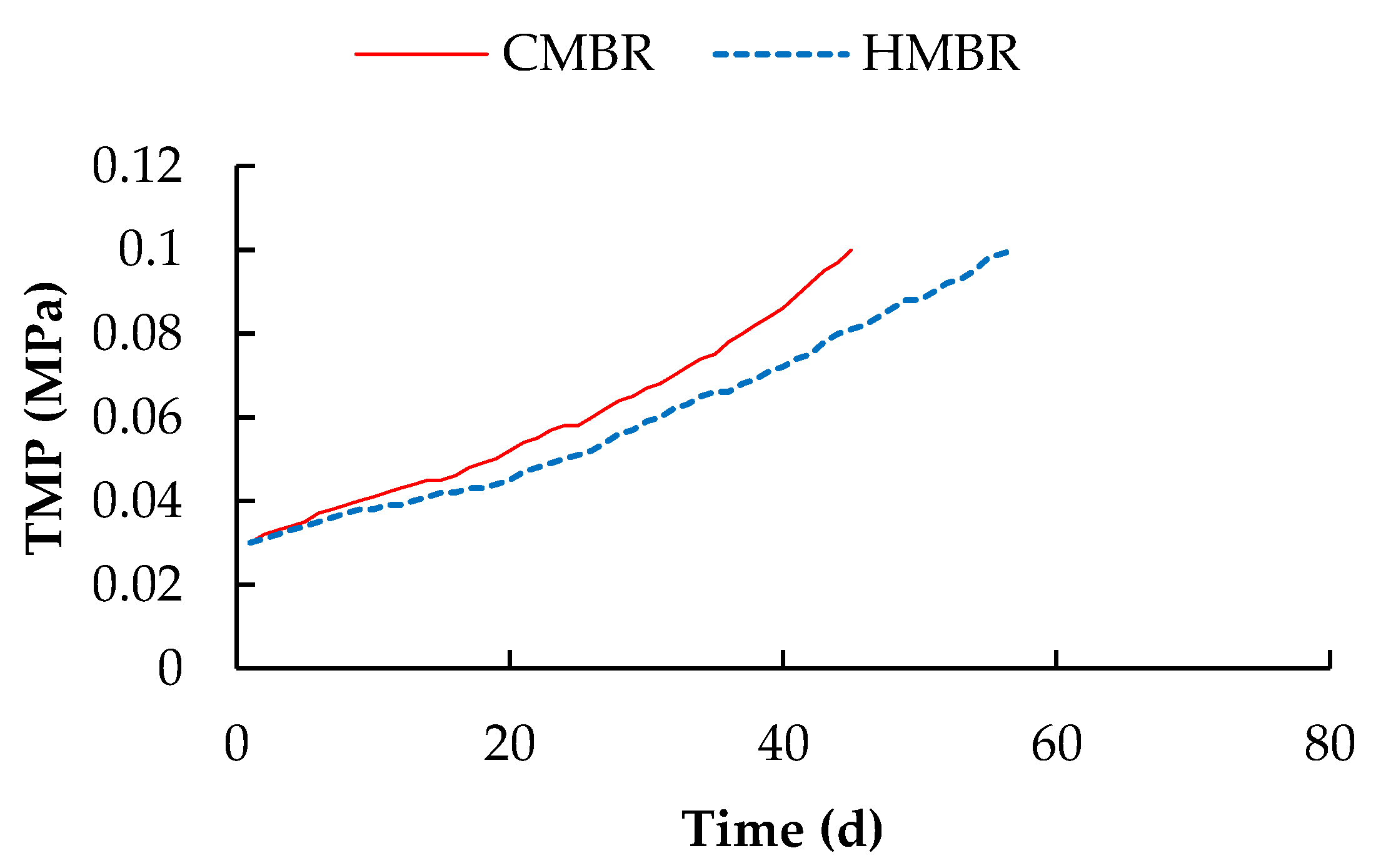
3.2. TMP Variation
3.3. EPS Distribution
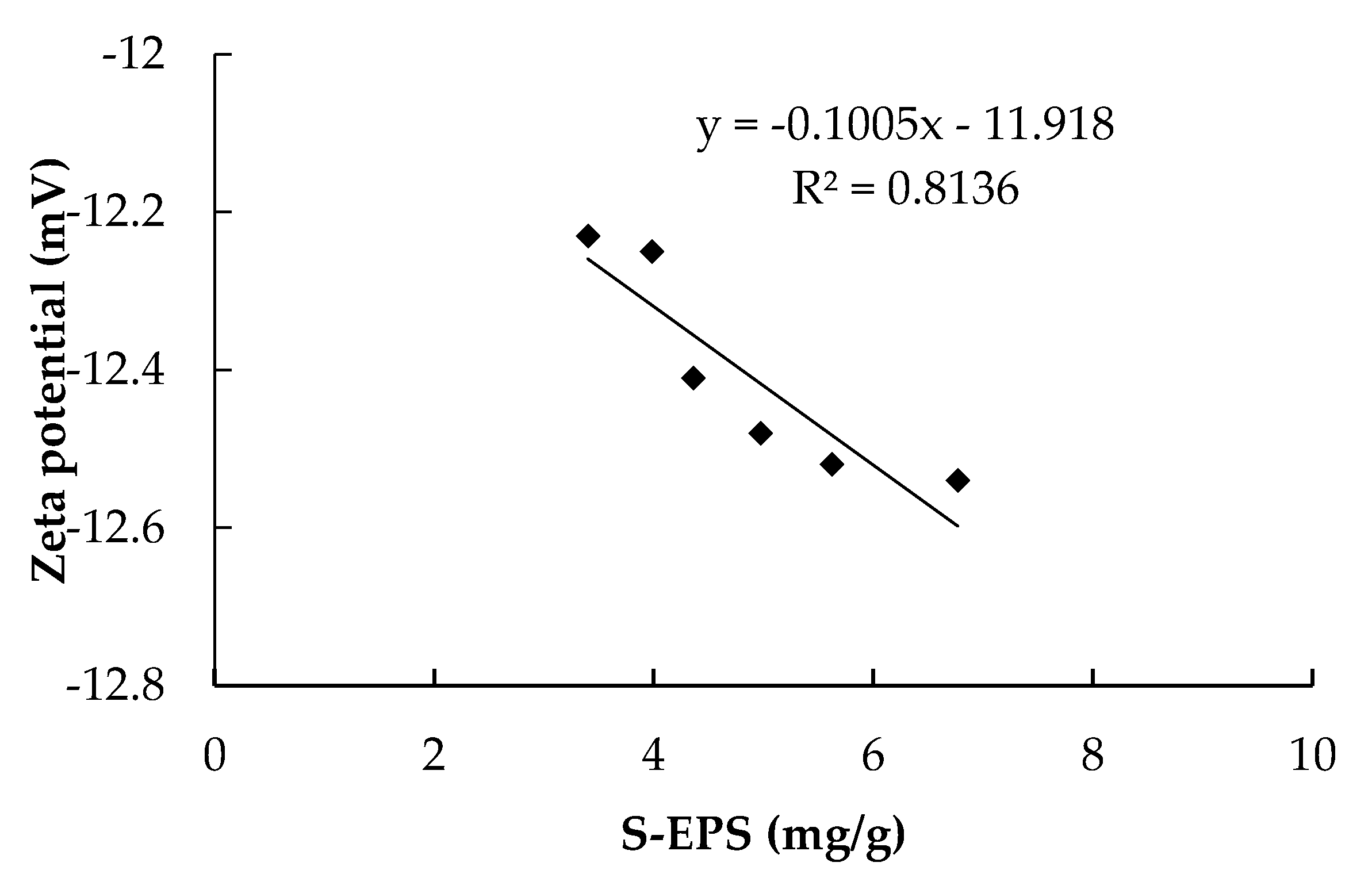
3.4. Relationship between EPS and Rc
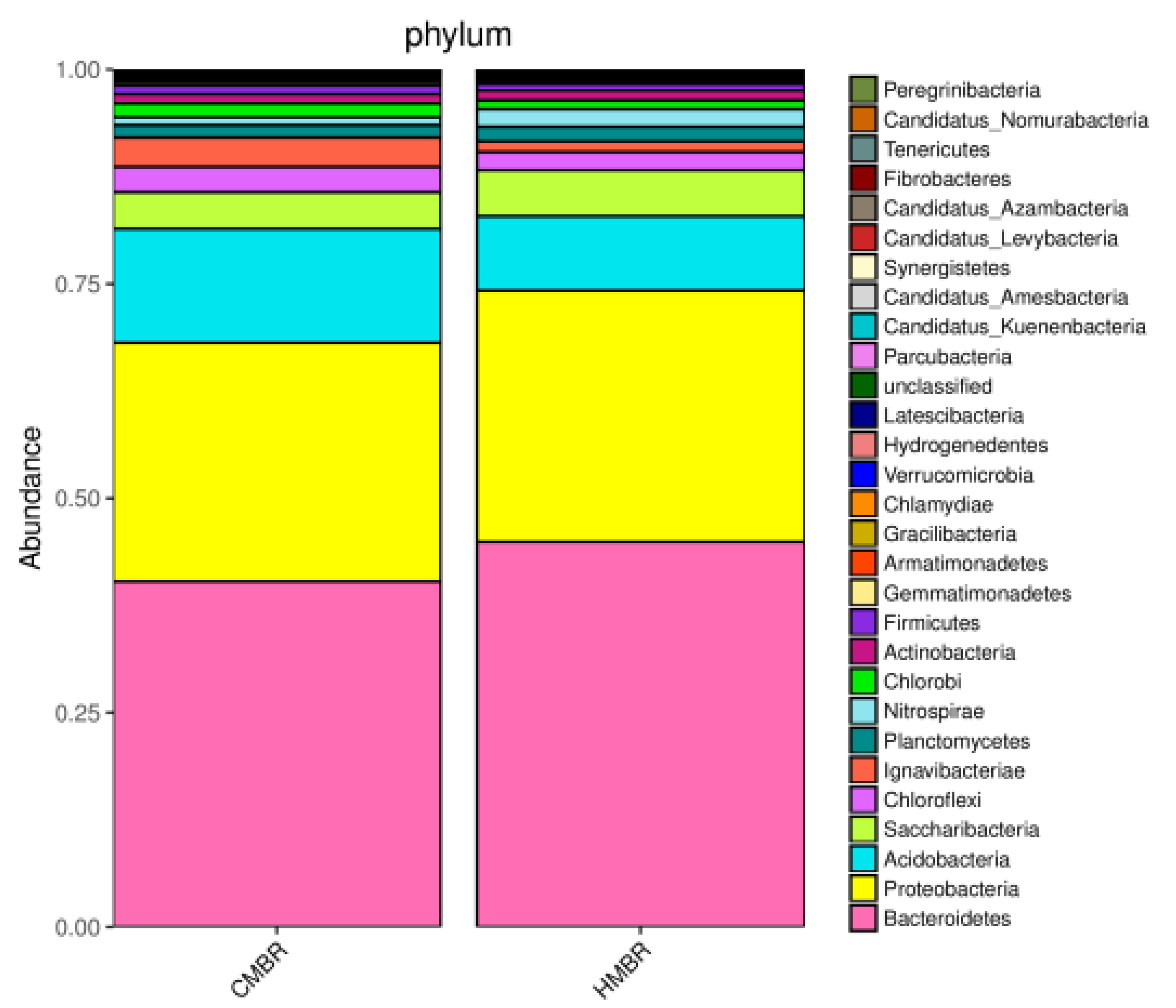
3.5. Microbial Community Structure
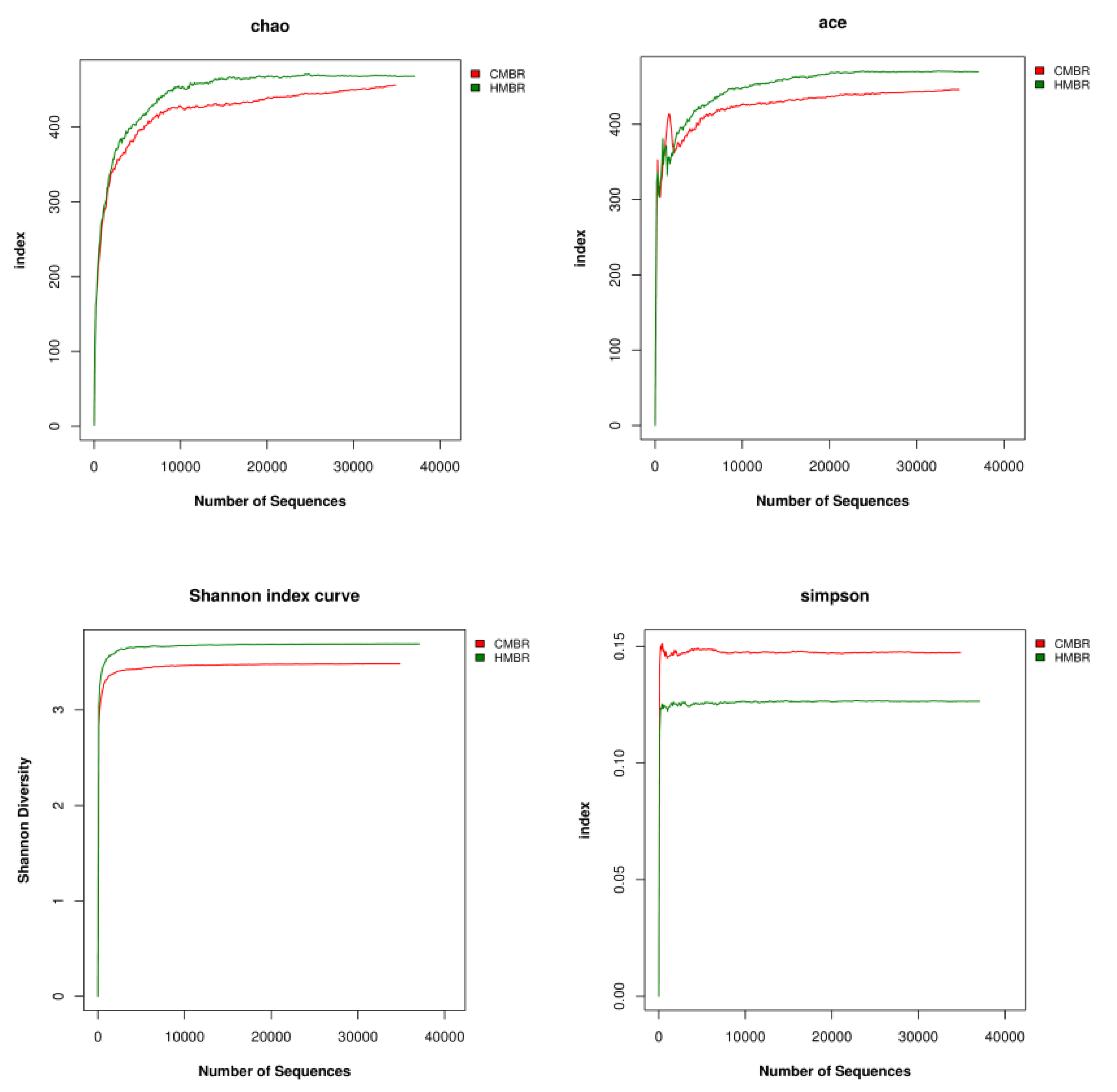
3.6. Alpha Diversity Analysis
3.7. Beta Diversity Analysis
3.8. Possible Mechanism of Membrane Fouling Control by HMBR
4. Conclusions
- Hybrid membrane bioreactor can remove organics and nutrients effectively. Averaged COD, BOD5, NH4+-N, TN and TP removals by which were higher by 4.1%, 4.2%, 1.0%, 17.7% and 2.2% respectively compared to those in the CMBR. Moreover, HMBR represents a good performance on membrane fouling control. When TMP reached 0.1 MPa, the membrane module in the HMBR operated for 57 days, which was longer by 26.7% compared to that in the CMBR.
- Soluble EPS in the HMBR was lower by 22.1% compared to the CMBR. Due to the S-EPS decrease, Zeta potential decreased, with activated sludge flocculability increasing and cake layer specific resistance decreasing accordingly. As a result, when the two reactors operated for the same time length of 45 days, Rp and Rc in the HMBR were lower by 19.0% and 25.2% respectively compared to those in the CMBR.
- A certain difference, albeit small, between the microbial community structures in the CMBR and HMBR was confirmed. Besides, both the species richness and diversity in the HMBR was apparently higher than those in the CMBR. This may be the direct reason why the HMBR can reduce the EPS effectively compared to the CMBR.
Author Contributions
Funding
Conflicts of Interest
References
- Brindle, K.; Stephensen, T. The application of membrane biological reactors for the treatment of wastewaters. Biotechnol. Bioeng. 2000, 49, 601–610. [Google Scholar] [CrossRef]
- Van Dijk, L.; Ronchen, G.C.G. Membrane bioreactors for wastewater treatment: The state of the art and new development. Water Sci. Technol. 1997, 35, 35–41. [Google Scholar] [CrossRef]
- Teychene, B.; Guigui, C.; Cabassud, C.; Amy, G. Toward a better identification of foulant species in MBR processes. Desalination 2008, 231, 27–34. [Google Scholar] [CrossRef]
- Drews, A. Membrane fouling in membrane bioreactors—Characterisation, contradictions, cause and cures. J. Membr. Sci. 2010, 363, 1–28. [Google Scholar] [CrossRef]
- Judd, S. The status of membrane bioreactor technology. Trends Biotechnol. 2008, 26, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Remmas, N.; Melidis, P.; Zerva, I.; Kristoffersen, J.B.; Nikolaki, S.; Tsiamis, G.; Ntougias, S. Dominance of candidate Saccharibacteria in a membrane bioreactor treating medium age landfill leachate: Effects of organic load on microbial communities, hydrolytic potential and extracellular polymeric substances. Bioresour. Technol. 2017, 238, 48–56. [Google Scholar] [CrossRef]
- Chang, I.S.; Clech, P.L.; Jefferson, B.; Judd, S. Membrane fouling in membrane bioreactors for wastewater treatment. J. Environ. Eng. 2002, 128, 1018–1029. [Google Scholar] [CrossRef]
- Chabalina, L.D.; Pastor, M.R.; Rico, D.P. Characterization of soluble and bound EPS obtained from 2 submerged membrane bioreactors by 3D-EEM and HPSEC. Talanta 2013, 115, 706–712. [Google Scholar] [CrossRef]
- Su, X.; Tian, Y.; Zuo, W.; Zhang, J.; Li, H.; Pan, X. Static adsorptive fouling of extracellular polymeric substances with different membrane materials. Water Res. 2014, 50, 267–277. [Google Scholar] [CrossRef]
- Comte, S.; Guibaud, G.; Baudu, M. Relations between extraction protocols for activated sludge extracellular polymeric substances (EPS) and EPS complexation properties Part 1 Comparison of the efficiency of eight EPS extraction methods. Enzyme Microb. Tech. 2006, 38, 237–245. [Google Scholar] [CrossRef]
- Poxon, T.L.; Darby, J.L. Extracellular polyanions in digested sludge: Measurement and relationship to sludge dewaterability. Water Res. 1997, 31, 749–758. [Google Scholar] [CrossRef]
- Sheng, G.-P.; Yu, H.-Q.; Li, X.-Y. Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: A review. Biotechnol. Adv. 2010, 28, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Laspidou, C.S.; Rittmann, B.E. A unified theory for extracellular polymeric substances, soluble microbial products, and active and inert biomass. Water Res. 2002, 36, 2711–2720. [Google Scholar] [CrossRef]
- Wan, C.-Y.; De Wever, H.; Diels, L.; Thoeye, C.; Liang, J.-B.; Huang, L.-N. Biodiversity and population dynamics of microorganisms in a full-scale membrane bioreactor for municipal wastewater treatment. Water Res. 2011, 45, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Guo, X.; Jiang, W.; Zhang, X.-X.; Wu, B. Mechanisms of microbial community structure and biofouling shifts under multivalent cations stress in membrane bioreactors. J. Hazard. Mater. 2017, 327, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.-C.; Lee, Y.-N.; Park, J.-S.; Lee, C.-H.; Lee, S.-H.; Chang, I.-S.; Ahn, T.-S. Correlation between dissolved oxygen concentration, microbial community and membrane permeability in a membrane bioreactor. Process. Biochem. 2006, 41, 1165–1172. [Google Scholar] [CrossRef]
- Falk, M.W.; Song, K.-G.; Matiasek, M.G.; Wuertz, S. Microbial community dynamics in replicate membrane bioreactors—Natural reproducible fluctuations. Water Res. 2009, 43, 842–852. [Google Scholar] [CrossRef]
- Stamper, D.M.; Walch, M.; Jacobs, R.N. Bacterial Population Changes in a Membrane Bioreactor for Graywater Treatment Monitored by Denaturing Gradient Gel Electrophoretic Analysis of 16S rRNA Gene Fragments. Appl. Environ. Microbiol. 2003, 69, 852–860. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Kim, S.; Yeon, K.-M.; Sang, B.-I.; Chun, J.; Lee, C.-H. Correlation between microbial community structure and biofouling in a laboratory scale membrane bioreactor with synthetic wastewater. Desalination 2012, 287, 209–215. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.C.; Liu, Y.; Yuan, H.; Du, Y. Performance of a hybrid membrane bioreactor in municipal wastewater treatment. Desalination 2010, 258, 143–147. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.C. Mechanism of nitrogen removal by a hybrid membrane reactor in municipal wastewater treatment. Desalin. Water Treat. 2014, 52, 5165–5171. [Google Scholar] [CrossRef]
- Higgins, M.J.; Novak, J.T. The effect of cations on the settling and dewatering of activated sludges: Laboratory results. Water Environ. Res. 1997, 69, 215–224. [Google Scholar] [CrossRef]
- Wang, X.C.; Liu, Q.; Liu, Y.J. Membrane Fouling Control of Hybrid Membrane Bioreactor: Effect of Extracellular Polymeric Substances. Sep. Sci. Technol. 2010, 45, 928–934. [Google Scholar] [CrossRef]
- Nagaoka, H.; Ueda, S.; Miya, A. Influence of bacterial extracellular polymers on the membrane separation activated sludge process. Water Sci. Tech. 1996, 34, 165–172. [Google Scholar] [CrossRef]
- Huang, X.; Gui, P.; Qian, Y. Effect of sludge retention time on microbial behaviour in a submerged membrane bioreactor. Process. Biochem. 2001, 36, 1001–1006. [Google Scholar] [CrossRef]
- APHA/AWWA/WEF. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association (APHA)/American Water Works Association (AWWA)/Water Environment Federation (WEF): Washington, DC, USA, 2012. [Google Scholar]
- Gaudy, A.F. Colorimetric determination of protein and carbohydrate. Ind. Water Wastes 1962, 7, 17–22. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Boil. Chem. 1951, 193, 265–275. [Google Scholar]
- Lee, J.; Ahn, W.-Y.; Lee, C.-H. Comparison of the filtration characteristics between attached and suspended growth microorganisms in submerged membrane bioreactor. Water Res. 2001, 35, 2435–2445. [Google Scholar] [CrossRef]
- Cho, J.; Ahn, K.H.; Lee, Y.H. Investigation of biological and fouling characteristics of submerged membrane bioreator process for wastewater treatment by model sensitivity analysis. Water Sci. Tech. 2004, 49, 245–254. [Google Scholar] [CrossRef]
- Chang, I.S.; Lee, C.H. Membrane filtration characteristics in membrane-coupled activated sludge system—The effect of physiological states of activated sludge on membrane fouling. Desalination 1998, 120, 221–233. [Google Scholar] [CrossRef]
- Rosenberger, S.; Laabs, C.; Lesjean, B.; Gnirss, R.; Amy, G.; Jekel, M.; Schrotter, J.-C. Impact of colloidal and soluble organic material on membrane performance in membrane bioreactors for municipal wastewater treatment. Water Res. 2006, 40, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Park, N.; Kwon, B.; Kim, I.S.; Cho, J. Biofouling potential of various NF membranes with respect to bacteria and their soluble microbial products (SMP): Characterizations, flux decline, and transport parameters. J. Membr. Sci. 2005, 258, 43–54. [Google Scholar] [CrossRef]
- Fang, H.H.; Jia, X.-S. Soluble microbial products (SMP) of acetotrophic methanogenesis. Bioresour. Technol. 1998, 66, 235–239. [Google Scholar] [CrossRef]
- Hughes, J.B.; Hellmann, J.J.; Richetts, T.H.; Bohannan, B.J.M. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl. Environ. Microbial. 2001, 67, 4399–4406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magurran, A.E. Measuring Biological Diversity; Blackwell Pusblishing: Oxford, UK, 2004. [Google Scholar]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, F.; Zhang, T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISMEJ 2014, 9, 683–695. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Description | Average |
|---|---|---|
| Chemical oxygen demand (COD) (mg/L) | 76.3–113.6 | 98.6 |
| Biological oxygen demand (BOD5) (mg/L) | 45.7–65.2 | 52.7 |
| Ammonia nitrogen (NH4+-N) (mg/L) | 26.3–34.8 | 29.2 |
| Total nitrogen (TN) (mg/L) | 31.4–52.6 | 37.4 |
| Total phosphorus (TP) (mg/L) | 2.47–3.74 | 3.20 |
| Temperature (°C) | 17.8–22.6 | 20.3 |
| pH | 7.53–7.69 | 7.61 |
| Parameter | CMBR | HMBR | ||
|---|---|---|---|---|
| Effluent (mg/L) | Averaged Removal (%) | Effluent (mg/L) | Averaged Removal (%) | |
| COD | 9.7–16.3 (13.7) | 86.1 | 6.7–13.1 (9.6) | 90.2 |
| BOD5 | 6.5–10.8 (8.9) | 83.1 | 4.8–9.4 (6.3) | 87.3 |
| NH4+-N | 0.3–1.1 (0.7) | 97.6 | 0.3–0.8 (0.4) | 98.6 |
| TN | 21.1–31.7 (25.8) | 31.0 | 17.2–25.1 (19.2) | 48.7 |
| TP | 0.53–1.06 (0.72) | 77.5 | 0.37–1.12 (0.65) | 79.7 |
| Operation Mode | Total Membrane Resistance (Rt) (/m) | Membrane Intrinsic Resistance (Rm) (/m) | Pore Blocking Resistance (Rp) (/m) | Cake Layer Resistance (Rc) (/m) |
|---|---|---|---|---|
| CMBR | 3.58 × 1013 | 0.71 × 1013 | 0.21 × 1013 | 2.70 × 1013 |
| HMBR | 2.90 × 1013 | 0.71 × 1013 | 0.17 × 1013 | 2.02 × 1013 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Yao, Y.; Xu, D. Mechanism of Membrane Fouling Control by HMBR: Effect of Microbial Community on EPS. Int. J. Environ. Res. Public Health 2020, 17, 1681. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph17051681
Liu Q, Yao Y, Xu D. Mechanism of Membrane Fouling Control by HMBR: Effect of Microbial Community on EPS. International Journal of Environmental Research and Public Health. 2020; 17(5):1681. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph17051681
Chicago/Turabian StyleLiu, Qiang, Ying Yao, and Delan Xu. 2020. "Mechanism of Membrane Fouling Control by HMBR: Effect of Microbial Community on EPS" International Journal of Environmental Research and Public Health 17, no. 5: 1681. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph17051681




