Diversity in the Enteric Viruses Detected in Outbreaks of Gastroenteritis from Mumbai, Western India
Abstract
:1. Introduction
2. Experimental Section
2.1. Epidemiological Investigation
2.2. Specimens
2.3. Laboratory Investigation
2.3.1. Nucleic Acid Extraction, PCR and RT-PCRs
2.3.2. Nucleotide Sequencing and Phylogentic Analyses
2.3.3. Nucleotide Sequence Accession Numbers
2.4. Environmental Investigation
3. Results
3.1. Epidemiological Investigation
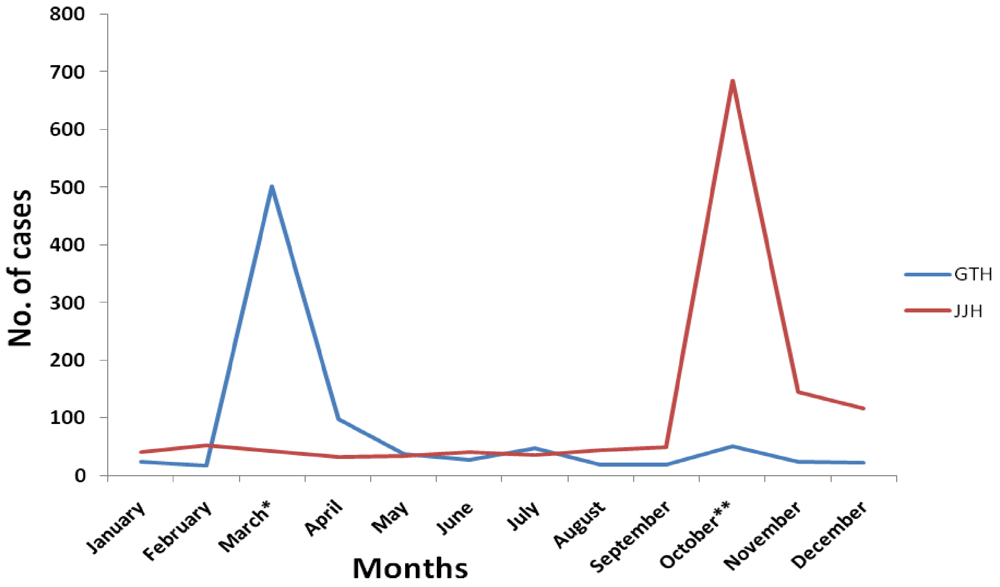
3.2. Enteric Virus Positivity
| Enteric virus | No. positive (%) | |
|---|---|---|
| March 2006 (n = 33) | October 2006 (n = 40) | |
| RVA | 11 (33.3) | 10 (25) |
| RVB | 4 (12.1) | 4 (10) |
| NoV | 4 (12.1) | 4 (10) |
| AdV | 2 (6.1) | 3 (7.5) |
| HAstV | 1 (3) | 0 |
| EV | 10 (30.3) | 16 (40) |
| AiV | 1 (3) | 3 (7.5) |
| Period of outbreak | RVA | RVB | NoV | AdV | HAstV | EV | |
|---|---|---|---|---|---|---|---|
| March, 2006 (n = 13) | M-689 | + | + | ||||
| M-791 | + | + | |||||
| M-633 | + | + | + | + | |||
| M-705 | + | + | |||||
| M-693 | + | + | |||||
| M-636 | + | + | |||||
| M-778 | + | + | |||||
| M-651 | + | + | |||||
| M-777 | + | + | |||||
| M-771 | + | + | |||||
| M-714 | + | + | + | ||||
| M-635 | + | + | |||||
| M-634 | + | + | |||||
| October, 2006 (n = 9) | M-836 | + | + | ||||
| M-859 | + | + | |||||
| M-029 | + | + | |||||
| M-038 | + | + | |||||
| M-308 | + | + | |||||
| M-864 | + | + | |||||
| M-857 | + | + | |||||
| M-361 | + | + | + | ||||
| M-364 | + | + |
3.3. Age Distribution
3.4. Nucleotide Sequencing and Phylogenetic Analyses
3.4.1. RVA
3.4.2. RVB
| Enteric Virus | Genotypes | March 2006 | October 2006 |
|---|---|---|---|
| RVA (n = 35) | G1P[8] | 0 | 2 |
| G2P[4] | 0 | 6 | |
| G9P[4] | 0 | 1 | |
| G12P[4] | 0 | 1 | |
| Only G typed | 0 | 11 | |
| Only P typed | 10 | 10 | |
| Both G and P nontypeable | 9 | 5 | |
| RVB (n = 11) | G2 | 7 | 4 |
| NoV (n = 10) | GII.2 | 2 | 0 |
| GII.4 | 3 | 2 | |
| GII.7 | 1 | 0 | |
| GII.12 | 0 | 1 | |
| GII.3 + GII.13 | 0 | 1 | |
| AdV (n = 16) | Type 12 | 2 | 0 |
| Type 31 | 0 | 1 | |
| Type 40 | 7 | 6 | |
| HAstV (n = 3) | HAstV-7 | 0 | 1 |
| HAstV-8 | 1 | 1 | |
| EV (n = 19) | EV-76 | 1 | 3 |
| EV-84 | 0 | 1 | |
| EV-89 | 0 | 1 | |
| EV-90 | 1 | 3 | |
| CA-13 | 1 | 0 | |
| CA-17 | 0 | 1 | |
| CA-19 | 1 | 0 | |
| CA-21 | 2 | 0 | |
| CA-19/22 | 1 | 0 | |
| Echo-21 | 0 | 1 | |
| Echo-32 | 0 | 2 | |
| AiV(n = 4) | Genotype B | 1 | 3 |
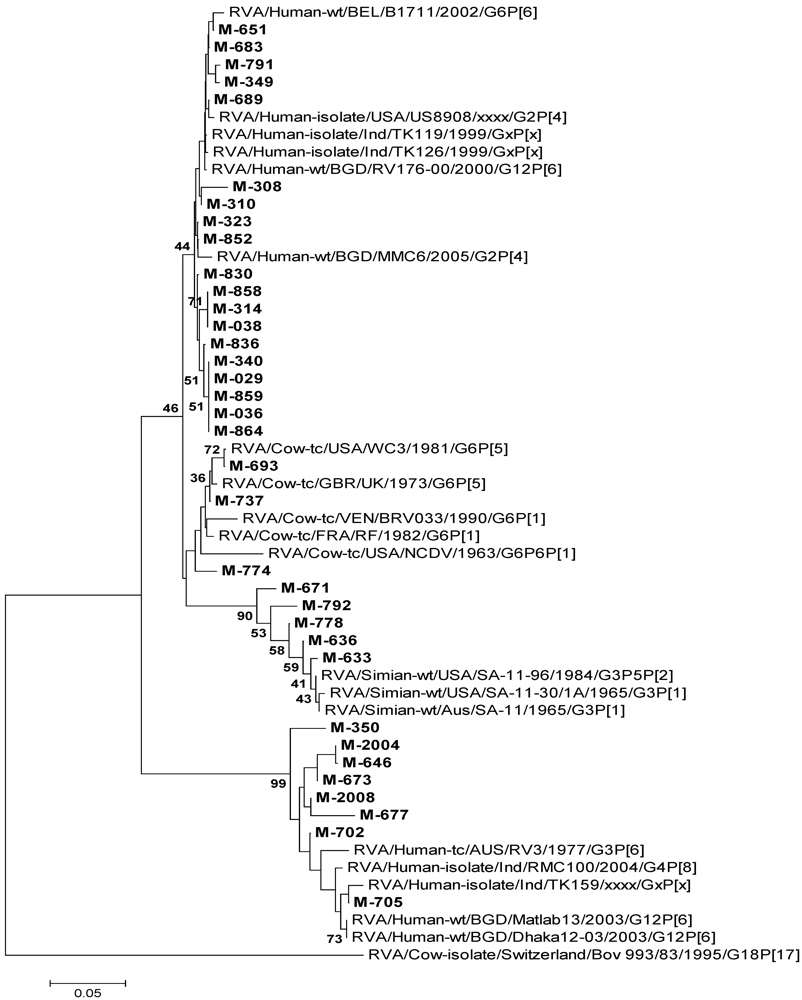
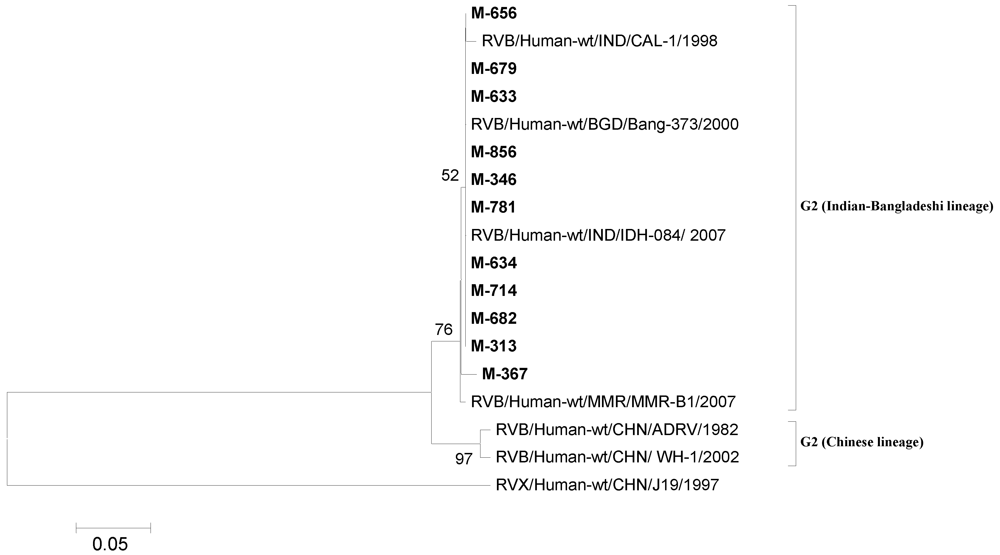
3.4.3. NoV
3.4.4. AdV


3.4.5. HAstV
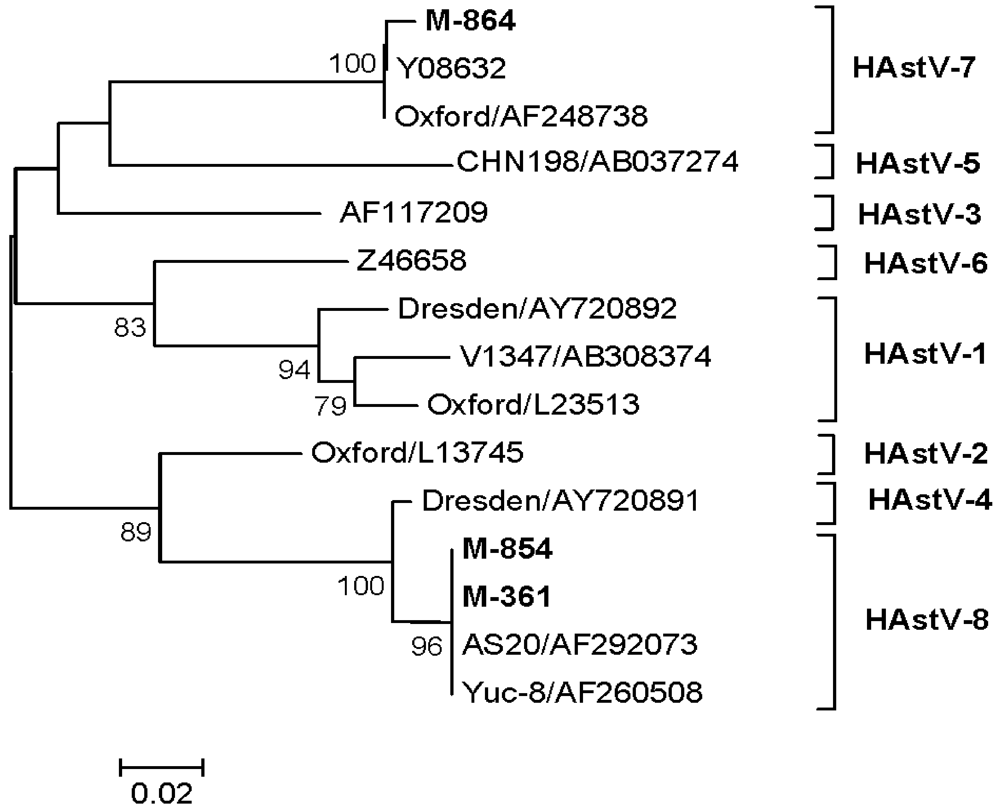
3.4.6. EV
3.4.7. AiV

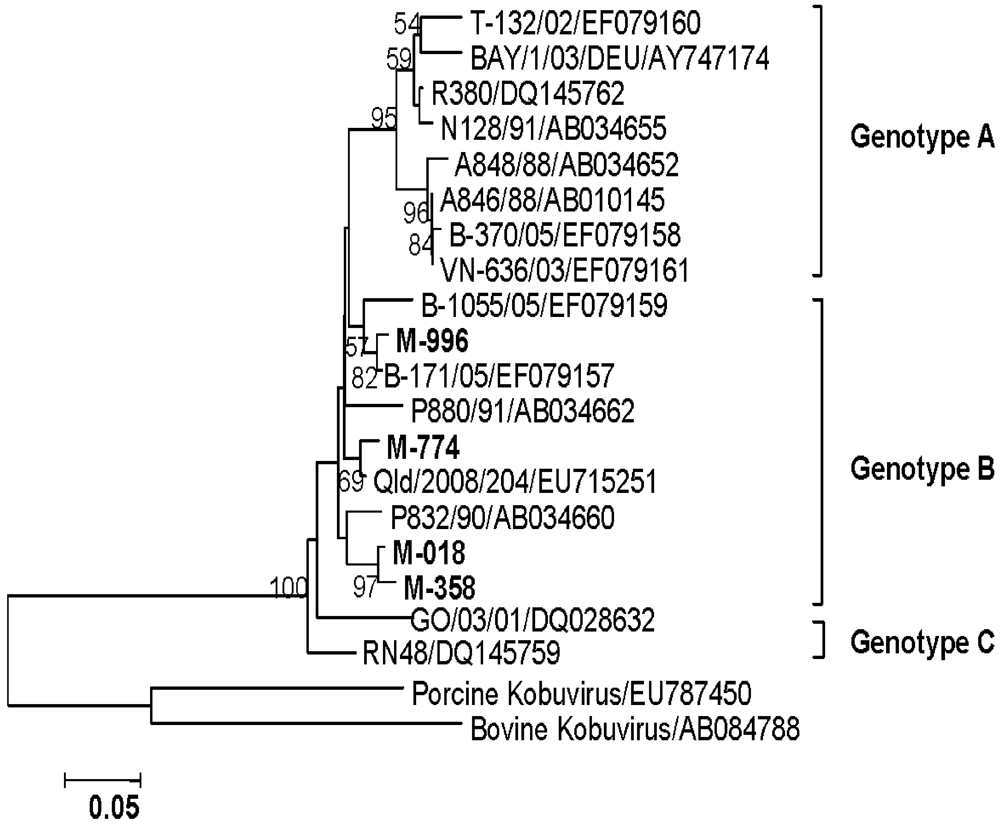
3.5. Environmental Investigation
4. Discussion
5. Conclusions
Acknowledgements
Conflict of Interest
References
- Navaneethan, U.; Giannella, R.A. Mechanisms of infectious diarrhea. Nat. Clin. Pract. Gastroenterol. Hepatol. 2008, 5, 637–647. [Google Scholar]
- Wilhelmi, I.; Roman, E.; Sánchez-Fauquier, A. Viruses causing gastroenteritis. Clin. Microbiol. Infect. 2003, 9, 247–262. [Google Scholar]
- Phan, T.G.; Nguyen, T.A.; Shimizu, H.; Yagyu, F.; Okitsu, S.; Müller, W.E.; Ushijima, H. Identification of enteroviral infection among infants and children admitted to hospital with acute gastroentritis in Ho Chi Minh City, Vietnam. J. Med. Virol. 2005, 77, 257–264. [Google Scholar]
- Shan, T.; Shan, T.; Wang, C.; Cui, L.; Yu, Y.; Delwart, E.; Zhao, W.; Zhu, C.; Lan, D.; Dai, X.; Hua, X. Picornavirus salivirus/klassevirus in children with diarrhea, China. Emerg. Infect. Dis. 2010, 16, 1303–1305. [Google Scholar]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Johne, R.; Kirkwood, C.D.; Martella, V.; Mertens, P.P.; Nakagomi, O.; Parreño, V.; Rahman, M.; Ruggeri, F.M.; Saif, L.J.; Santos, N.; Steyer, A.; Taniguchi, K.; Patton, J.T.; Desselberger, U.; Van Ranst, M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar]
- Barman, P.; Ghosh, S.; Samajdar, S.; Mitra, U.; Dutta, P.; Bhattacharya, S.K.; Krishnan, T.; Kobayashi, N.; Naik, T.N. RT-PCR based diagnosis revealed importance of human group B rotavirus infection in childhood diarrhoea. J. Clin. Virol. 2006, 36, 222–227. [Google Scholar]
- Aung, T.S.; Kobayashi, N.; Nagashima, S.; Ghosh, S.; Aung, M.S.; Oo, K.Y.; Win, N. Detection of group B rotavirus in an adult with acute gastroenteritis in Yangon, Myanmar. J. Med. Virol. 2009, 81, 1968–1974. [Google Scholar]
- Hall, A.J.; Vinjé, J.; Lopman, B.; Park, G.W.; Yen, C.; Gregoricus, N.; Parashar, U. Updated norovirus outbreak management and disease prevention guidelines. MMWR Morb. Mortal. Wkly. Rep. 2011, 60, 1–15. [Google Scholar]
- Brown, M. Laboratory identification of adenoviruses associated with gastroenteritis in Canada from 1983 to 1986. J. Clin. Microbiol. 1990, 28, 1525–1529. [Google Scholar]
- Verma, H.; Chitambar, S.D.; Gopalkrishna, V. Identification and characterization of enteric adenoviruses in infants and children hospitalized for acute gastroenteritis. J. Med. Virol. 2009, 81, 60–64. [Google Scholar]
- Mendez, E.; Arias, C.F. Astroviruses. In Fields Virology, 5th; Fields, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Monath, T.P., Melnick, J.L., Roizman, B., Straus, S.E., Eds.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 981–1000. [Google Scholar]
- Oberste, M.S.; Maher, K.; Nix, W.A.; Michele, S.M.; Uddin, M.; Schnurr, D.; Al-Busaidy, S.; Akoua-Koffi, C.; Pallansch, M.A. Molecular identification of 13 new enterovirus types, EV79–88, EV97, and EV100–101, members of the species human enterovirus B. Virus Res. 2007, 128, 34–42. [Google Scholar] [CrossRef]
- Ambert-Balay, K.; Lorrot, M.; Bon, F.; Giraudon, H.; Kaplon, J.; Wolfer, M.; Lebon, P.; Gendrel, D.; Pothier, P. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J. Clin. Microbiol. 2008, 46, 1252–1258. [Google Scholar]
- Bhattacharya, R.; Sahoo, G.C.; Nayak, M.K.; Ghosh, S.; Dutta, P.; Bhattacharya, M.K.; Mitra, U.; Gangopadhyay, D.; Dutta, S.; Niyogi, S.K.; Saha, D.R.; Naik, T.N.; Bhattacharya, S.K.; Krishnan, T. Molecular epidemiology of human astrovirus infections in Kolkata, India. Infect. Genet. Evol. 2006, 6, 425–435. [Google Scholar]
- Chhabra, P.; Dhongade, R.K.; Kalrao, V.R.; Bavdekar, A.R.; Chitambar, S.D. Epidemiological, clinical and molecular features of norovirus infections in western India. J. Med. Virol. 2009, 81, 922–932. [Google Scholar]
- Kang, G.; Arora, R.; Chitambar, S.; Deshpande, J.; Gupta, M.D.; Kulkarni, M.; Naik, T.N.; Mukherji, D.; Venkatasubramanium, S.; Gentsch, J.R.; Glass, R.I.; Parashar, U.D. Multicenter, hospital-based surveillance of rotavirus disease and strains among Indian children. J. Infect. Dis. 2009, 200, S147–S153. [Google Scholar]
- Tatte, V.S.; Gentsch, J.R.; Chitambar, S.D. Characterization of group A rotavirus infections in adolescents and adults from Pune, India: 1993-1996 and 2004-2007. J. Med. Virol. 2010, 82, 519–527. [Google Scholar]
- Verma, H.; Chitambar, S.D.; Gopalkrishna, V. Astrovirus associated acute gastroenteritis in western India: predominance of dual serotype strains. Infect. Genet. Evol. 2010, 10, 575–579. [Google Scholar]
- Mumbai—Wikipedia, the Free Encyclopedia. Available online: http://en.wikipedia.org/wiki/Mumbai (accessed on 9 July 2011).
- Wilde, J.; Eiden, J.; Yolken, R. Removal of inhibitory substances from human faecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reaction. J. Clin. Microbiol. 1990, 28, 1300–1307. [Google Scholar]
- Gentsch, J.R.; Glass, R.I.; Woods, P.; Gouvea, V.; Gorziglia, M.; Flores, J.; Das, B.K.; Bhan, M.K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1365–1373. [Google Scholar]
- Freeman, M.M.; Kerin, T.; Jennifer, H.; Caustland, K.M.; Gentsch, J. Enhancement of detection and quantification of rotavirus in stool using a modified real time RT-PCR assay. J. Med. Virol. 2008, 80, 1489–1496. [Google Scholar]
- Ando, T.; Monroe, S.S.; Gentsch, J.R.; Jin, Q.; Lewis, D.C.; Glass, R.I. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and southern hybridization. J. Clin. Microbiol. 1995, 33, 64–71. [Google Scholar]
- Girish, R.; Broor, S.; Dar, L.; Ghosh, D. Foodborne outbreak caused by a Norwalk- like virus in India. J. Med. Virol. 2002, 67, 603–607. [Google Scholar]
- Allard, A.; Girones, R.; Juto, P.; Wadell, G. Polymerase chain reaction for detection of adenoviruses in stool samples. J. Clin. Microbiol. 1990, 28, 2659–2667. [Google Scholar]
- Noel, J.S.; Lee, T.W.; Kurtz, J.B.; Glass, R.I.; Monroe, S.S. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 1995, 33, 797–801. [Google Scholar]
- Sapkal, G.N.; Bondre, V.P.; Fulmali, P.V.; Patil, P.; Gopalkrishna, V.; Dadhania, V.; Ayachit, V.M.; Gangale, D.; Kushwaha, K.P.; Rathi, A.K.; Chitambar, S.D.; Mishra, A.C.; Gore, M.M. Enteroviruses in patients with acute encephalitis, Uttar Pradesh, India. Emerg. Infect. Dis. 2009, 15, 295–298. [Google Scholar]
- Yamashita, T.; Sugiyama, M.; Tsuzuki, H.; Sakae, K.; Suzuki, Y.; Miyazaki, Y. Application of a reverse transcription-PCR for identification and differentiation of aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 2000, 38, 2955–2961. [Google Scholar]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods. 2002, 100, 107–114. [Google Scholar]
- Oberste, M.S.; Maher, K.; Kilpatrick, D.R.; Flemister, M.R.; Brown, B.A.; Pallansch, M.A. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 1999, 37, 1288–1293. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Arankalle, V.A.; Sarada, D.K.L.; Lole, K.S.; Shenoy, K.T.; Verma, V.; Haneephabi, M. Molecular characterization of hepatitis A virus from a large outbreak from Kerala, India. Indian. J. Med. Res. 2006, 123, 760–769. [Google Scholar]
- Gallimore, C.I.; Pipkin, C.; Shrimpton, H.; Green, A.D.; Pickford, Y.; McCartney, C.; Sutherland, G.; Brown, D.W.G.; Gray, J.J. Detection of multiple enteric virus strains within a food borne outbreak of gastroenteritis: an indication of the source of contamination. Epidemiol. Infect. 2005, 133, 41–47. [Google Scholar]
- Kittigul, L.; Pombubpa, K.; Taweekate, Y.; Yeephoo, T.; Khamrin, P.; Ushijima, H. Molecular characterization of rotaviruses, noroviruses, sapoviruses, and adenoviruses in patients with acute gastroenteritis in Thailand. J. Med. Virol. 2009, 81, 345–353. [Google Scholar]
- Patel, J.R.; Daniel, J.; Mathan, V.I. An epidemic of acute diarrhoea in rural southern India associated with echovirus type 11 infection. J. Hyg. 1985, 95, 483–492. [Google Scholar]
- Kelkar, S.D.; Ray, P.G.; Shinde, D.N. An epidemic of rotavirus diarrhoea in Jawhar Taluk, Thane District, Maharashtra, India, December 2000-January 2001. Epidemiol. Infect. 2004, 132, 337–341. [Google Scholar]
- Chitambar, S.D.; Lahon, A.; Tatte, V.S.; Maniya, N.H.; Tambe, G.U.; Khatri, K.I.; Desai, H.S.; Ugare, M.R.; Kulkarni, S.V.; Waghmare, A.P. Occurrence of group B rotavirus infections in the outbreaks of acute gastroenteritis from western India. Ind. J. Med. Res. 2011, 134, 399–400. [Google Scholar]
- Villena, C.; Gabrieli, R.; Pinto, R.M.; Guix, S.; Donia, D.; Buonuomo, E.; Palombi, L.; Cenko, F.; Bino, S.; Bosch, A.; Divizia, M.A. Large infantile gastroenteritis outbreak in Albania caused by multiple emerging rotavirus genotypes. Epidemiol. Infect. 2003, 131, 1105–1110. [Google Scholar]
- Iijima, Y.; Iwamoto, T.; Nukuzuma, S.; Ohishi, H.; Hayashi, K.; Kobayashi, N. An outbreak of rotavirus infection among adults in an institution for rehabilitation: long-term residence in a closed community as a risk factor for rotavirus illness. Scand. J. Infect. Dis. 2006, 38, 490–496. [Google Scholar]
- Räsänen, S.; Lappalainen, S.; Kaikkonen, S.; Hämäläinen, M.; Salminen, M.; Vesikari, T. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 2010, 138, 1227–1234. [Google Scholar]
- Fukuda, S.; Sasaki, Y.; Takao, S.; Seno, M. Recombinant norovirus implicated in gastroenteritis outbreaks in Hiroshima prefecture, Japan. J. Med. Virol. 2008, 80, 921–928. [Google Scholar]
- Grohmann, G.S.; Glass, R.I.; Pereira, H.G.; Monroe, S.S.; Hightower, A.W.; Weber, R.; Bryan, R.T. Enteric viruses and diarrhea in HIV-infected patients. N. Engl. J. Med. 1993, 329, 14–20. [Google Scholar]
- One More Succumbs to Gastroenteritis in Mumbai, More People Hospitalized. Available online: http://www.medindia.net/news/view_news_main.asp?x=8583 (accessed on 17 March 2006).
- Malaria, Dengue and Now Cholera? Available online: http://www.dnaindia.com/mumbai/report_malaria-dengue-and-now-cholera_1058561 (accessed on 15 October 2006).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chitambar, S.; Gopalkrishna, V.; Chhabra, P.; Patil, P.; Verma, H.; Lahon, A.; Arora, R.; Tatte, V.; Ranshing, S.; Dhale, G.; et al. Diversity in the Enteric Viruses Detected in Outbreaks of Gastroenteritis from Mumbai, Western India. Int. J. Environ. Res. Public Health 2012, 9, 895-915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph9030895
Chitambar S, Gopalkrishna V, Chhabra P, Patil P, Verma H, Lahon A, Arora R, Tatte V, Ranshing S, Dhale G, et al. Diversity in the Enteric Viruses Detected in Outbreaks of Gastroenteritis from Mumbai, Western India. International Journal of Environmental Research and Public Health. 2012; 9(3):895-915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph9030895
Chicago/Turabian StyleChitambar, Shobha, Varanasi Gopalkrishna, Preeti Chhabra, Pooja Patil, Harsha Verma, Anismrita Lahon, Ritu Arora, Vaishali Tatte, Sujata Ranshing, Ganesh Dhale, and et al. 2012. "Diversity in the Enteric Viruses Detected in Outbreaks of Gastroenteritis from Mumbai, Western India" International Journal of Environmental Research and Public Health 9, no. 3: 895-915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph9030895




