Quantitative Spectral Data Analysis Using Extreme Learning Machines Algorithm Incorporated with PCA
Abstract
:1. Introduction
2. Materials and Methods
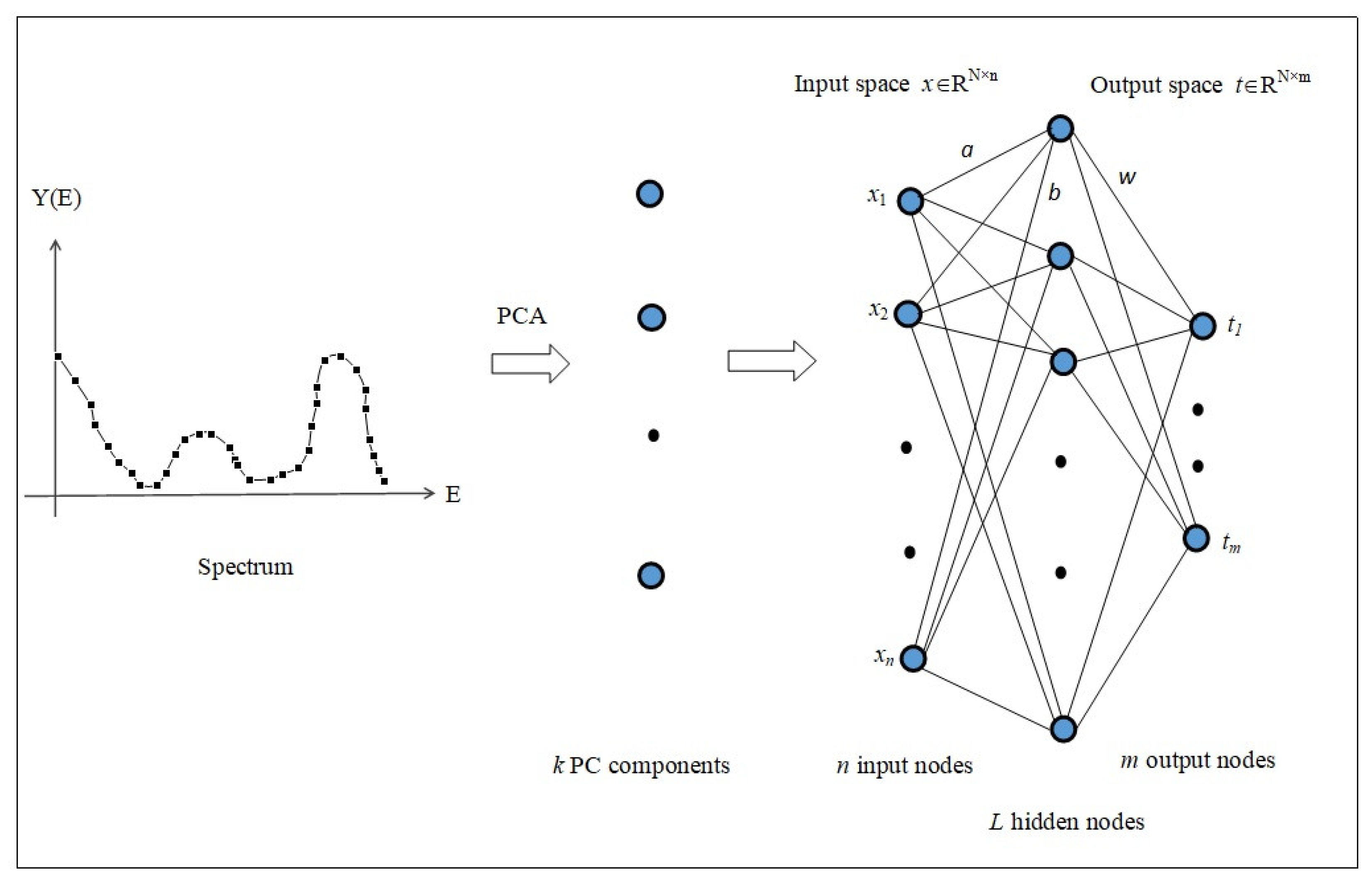
2.1. Spectral Data Analysis and Multivariate Regression Problems
2.2. Solution by an Enhanced ELM
2.3. Principal Component Analysis and Dimensionality Reduction
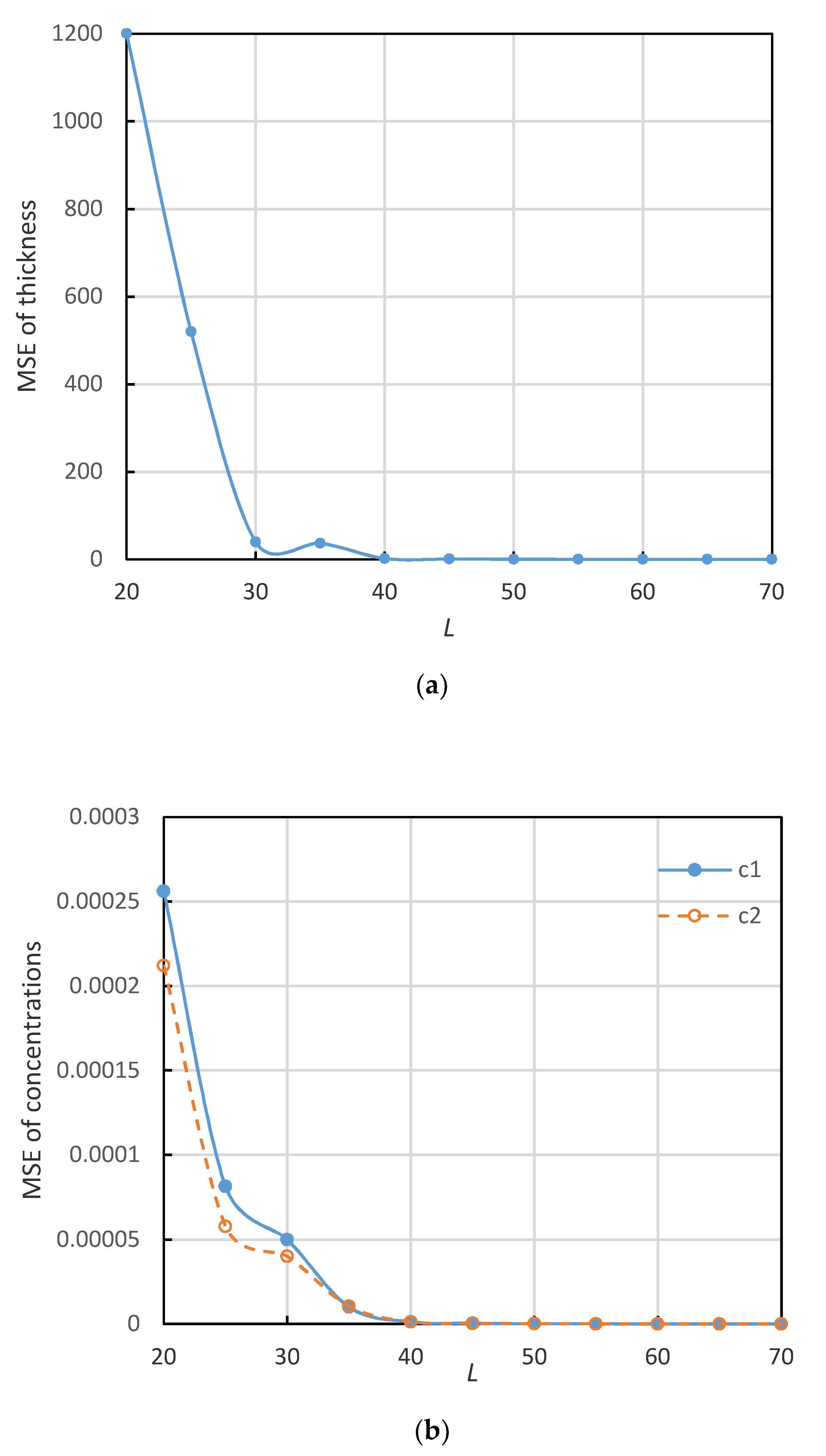
3. Experiment
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barradas, N.P.; Jeynes, C.; Webber, R.; Keissig, U.; Grotzschel, R. Unambiguous automatic evaluation of multiple ion beam analysis data with simulated annealing. Nucl. Instr. Meth. Phys. Res. B 1999, 149, 233–238. [Google Scholar] [CrossRef]
- van Laarhoven, P.J.M.; Aarts, E.H.L. Simulated Annealing. In Simulated Annealing: Theory and Applications. Mathematics and Its Applications; Springer: Berlin/Heidelberg, Germany, 1987; Volume 37. [Google Scholar]
- Orosz, J.; Jacobson, S. Analysis of Static Simulated Annealing Algorithms. J. Optim. Theory Appl. 2002, 115, 165–182. [Google Scholar] [CrossRef]
- Siddique, N.; Adeli, H. Simulated Annealing, Its Variants and Engineering Applications. Int. J. Artif. Intell. Tools 2016, 25, 1630001–1630025. [Google Scholar] [CrossRef]
- Barradas, N.; Vieira, A. Artificial neural network algorithm for analysis of Rutherford backscattering data. Phys. Rev. E 2000, 62, 5818–5829. [Google Scholar] [CrossRef] [PubMed]
- Demeulemeester, J.; Smeets, D.; Barradas, N.; Vieira, A.; Comrie, C.; Temst, K.; Vantomme, A. Artificial neural networks for instantaneous analysis of real-time Rutherford backscattering spectra. Nucl. Instr. Meth. Phys. Res. Sect. B Beam Interact. Mater. Atoms 2010, 268, 1676–1681. [Google Scholar] [CrossRef]
- Nene, N.R.; Vieira, A.; Barradas, N.P. Artificial neural networks analysis of RBS and ERDA spectra of multilayered multi-elemental samples. Nucl. Instr. Meth. Phys. Res. B 2006, 246, 471–478. [Google Scholar] [CrossRef]
- Wang, J.-S.; Chen, Y.-P. A fully automated recurrent neural network for unknown dynamic system identification and control. IEEE Trans. Circuits Syst. I Regul. Pap. 2006, 53, 1363–1372. [Google Scholar] [CrossRef]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Lam, L.H.T.; Le, N.H.; Van Tuan, L.; Ban, H.T.; Hung, T.N.K.; Nguyen, N.T.K.; Dang, L.H.; Le, N.Q.K. Machine Learning Model for Identifying Antioxidant Proteins Using Features Calculated from Primary Sequences. Biology 2020, 9, 325. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Do, D.T.; Chiu, F.-Y.; Yapp, E.K.Y.; Yeh, H.-Y.; Chen, C.-Y. XGBoost Improves Classification of MGMT Promoter Methylation Status in IDH1 Wildtype Glioblastoma. J. Pers. Med. 2020, 10, 128. [Google Scholar] [CrossRef]
- Deist, T.M.; Dankers, F.J.W.M.; Valdes, G.; Wijsman, R.; Hsu, I.; Oberije, C.; Lustberg, T.; Van Soest, J.; Hoebers, F.; Jochems, A.; et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys. 2018, 45, 3449–3459. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Kasun, L.L.C.; Zhou, H.; Huang, G.B. Representational learning with ELMs for big data. IEEE Intell. Syst. 2013, 28, 31–34. [Google Scholar]
- Huang, G.-B. An Insight into Extreme Learning Machines: Random Neurons, Random Features and Kernels. Cogn. Comput. 2014, 6, 376–390. [Google Scholar] [CrossRef]
- Han, H.-G.; Wang, L.-D.; Qiao, J. Hierarchical extreme learning machine for feedforward neural network. Neurocomputing 2014, 128, 128–135. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhou, H.; Ding, X.; Zhang, R. Extreme Learning Machine for Regression and Multiclass Classification. IEEE Trans. Syst. Man Cybern. Part B 2012, 42, 513–529. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Huang, G.B.; Song, S.; You, K. Trends in extreme learning machines: A review. Neural Netw. 2015, 61, 32–48. [Google Scholar] [CrossRef]
- Zheng, W.; Fu, X.; Ying, Y. Spectroscopy-based food classification with extreme learning machine. Chemom. Intell. Lab. Syst. 2014, 139, 42–47. [Google Scholar] [CrossRef]
- Zheng, W.; Shu, H.; Tang, H.; Zhang, H. Spectra data classification with kernel extreme learning machine. Chemom. Intell. Lab. Syst. 2019, 192. [Google Scholar] [CrossRef]
- da Costa, N.L.; Llobodanin, L.A.G.; de Lima, M.D.; Castro, I.A.; Barosa, R. Geographical recognition of Syrah wines by combining feature selection with extreme learning machine. Measurement 2018, 120, 92–99. [Google Scholar] [CrossRef]
- Quadram Institute. Available online: https://csr.quadram.ac.uk/example-datasets-for-download/ (accessed on 5 March 2020).
- Khan, B.; Wang, Z.; Han, F.; Iqbal, A.; Masood, R.J. Fabric weave pattern and yarn colour recognition and classification using a deep ELM network. Algorithms 2017, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Song, G.; Dai, Q.; Han, X.; Guo, L. Two novel ELM-based stacking deep models focused on image recognition. Appl. Intell. 2020, 50, 1345–1366. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, G.-B.; Lin, Z.; Wang, H.; Soh, Y.C. Stacked Extreme Learning Machines. IEEE Trans. Cybern. 2015, 45, 2013–2025. [Google Scholar] [CrossRef]
- Tarantola, A. Inverse problem theory and methods for model parameter estimation. SIAM 2004. [Google Scholar] [CrossRef] [Green Version]
- Mosegaard, K.; Sambridge, M. Monte Carlo analysis of inverse problems. Inverse Probl. 2002, 18, R29–R54. [Google Scholar] [CrossRef]
- Kotai, E. Computer methods for analysis and simulation of RBS and ERDA spectra. Nucl. Instr. Meth. Phys. Res. B 1994, 85, 588–596. [Google Scholar] [CrossRef]
- Denison, D.G.T.; Mallick, B.K.; Smith, A.F.M. Automatic Bayesian curve fitting. J. R. Stat. Soc. Ser. B 1998, 60, 333–350. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer Series in Statistics; Springer: New York, NY, USA, 2009. [Google Scholar]
- Broomhead, D.S.; Lowe, D. Multivariable function interpolation and adaptive networks. Complex Syst. 1998, 2, 321–355. [Google Scholar]
- Park, J.; Sandberg, I.W. Universal Approximation Using Radial-Basis-Function Networks. Neural Comput. 1991, 3, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, L.D. A Novel Method of Curve Fitting Based on Optimized Extreme Learning Machine. Appl. Artif. Intell. 2020, 34, 849–865. [Google Scholar] [CrossRef]
- Li, M.; Wibowo, S.; Guo, W. Nonlinear Curve Fitting Using Extreme Learning Machines and Radial Basis Function Networks. Comput. Sci. Eng. 2019, 21, 6–15. [Google Scholar] [CrossRef]
- Serre, D. Matrices: Theory and Applications; Springer: New York, NY, USA, 2002. [Google Scholar]
- Haykin, S. Neural Networks: A Comprehensive Foundation, 3rd ed.; Pearson: London, UK, 2009. [Google Scholar]
- Serneels, S.; Verdonck, T. Principal component analysis for data containing outliers and missing elements. Comput. Stat. Data Anal. 2008, 52, 1712–1727. [Google Scholar] [CrossRef]
- Belsley, D.A.; Kuh, E.; Welsch, R.E. Regression Diagnostics. Identifying Influential Data and Sources of Collinearity; Wiley: New York, NY, USA, 1980. [Google Scholar]
- Mayer, M. SIMNRA 7.02 User’s Guide. Max-Planck Institute of Plasma Physics; Max-Planck-Institut für Plasmaphysik: Garching, Germany, 2019. [Google Scholar]
- Mayer, M. Improved Physics in SIMNRA 7. Nucl. Instr. Meth. Phys. Res. B 2014, 332, 176–187. [Google Scholar] [CrossRef]
- Extreme Learning Machines (ELM). Extreme Learning Machines (ELM): Filling the Gap between Frank Rosenblatt’s Dream and John von Neumann’s Puzzle. Available online: https://www.ntu.edu.sg/home/egbhuang/ (accessed on 5 March 2020).
- Xie, Z.; Xu, K.; Shan, W.; Liu, L.; Xiong, Y.; Huang, H. Projective Feature Learning for 3D Shapes with Multi-View Depth Images. Comput. Graph. Forum 2015, 34, 1–11. [Google Scholar] [CrossRef]
- Aziz, S.; Mohamed, E.A.; Youssef, F. Traffic Sign Recognition Based On Multi-feature Fusion and ELM Classifier. Procedia Comput. Sci. 2018, 127, 146–153. [Google Scholar] [CrossRef]




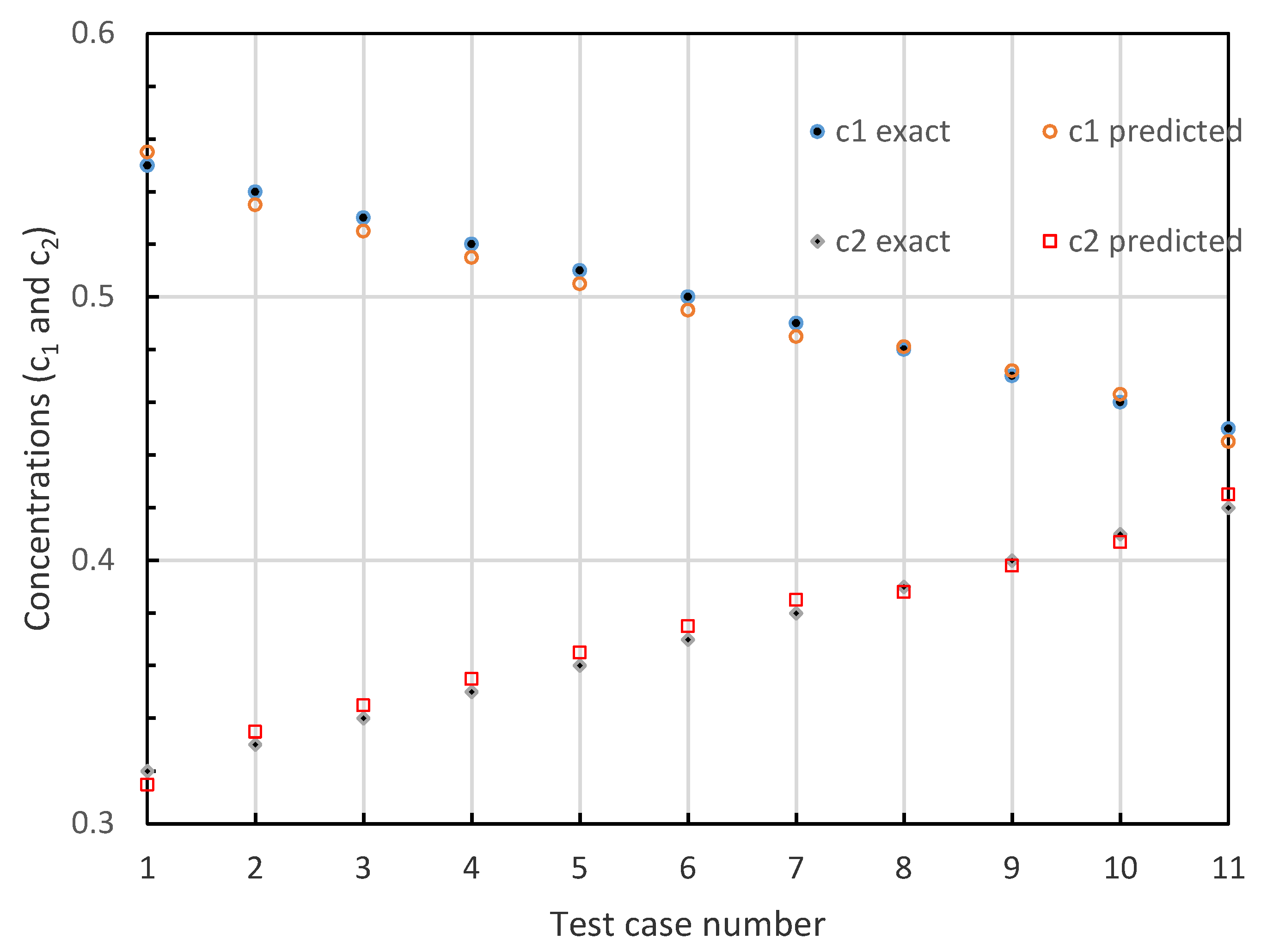
| Sample No | Exact Values | ELM Predictions | ||||
|---|---|---|---|---|---|---|
| t × 1015 at/cm2 | c1 | c2 | t × 1015 at/cm2 | c1 | c2 | |
| 1 | 697.82 | 0.5400 | 0.3300 | 704.37 | 0.5389 | 0.3311 |
| 2 | 1295.88 | 0.5100 | 0.3600 | 1302.48 | 0.5111 | 0.3589 |
| 3 | 2397.33 | 0.4800 | 0.3900 | 2413.15 | 0.4762 | 0.3938 |
| 4 | 3072.98 | 0.4500 | 0.4200 | 3065.29 | 0.4484 | 0.4216 |
| Sample No | Exact Values | MLP Predictions | ||||
|---|---|---|---|---|---|---|
| t × 1015 at/cm2 | c1 | c2 | t × 1015 at/cm2 | c1 | c2 | |
| 1 | 697.82 | 0.5400 | 0.3300 | 708.55 | 0.5366 | 0.3334 |
| 2 | 1295.88 | 0.5100 | 0.3600 | 1290.33 | 0.5134 | 0.3566 |
| 3 | 2397.33 | 0.4800 | 0.3900 | 2422.32 | 0.4701 | 0.3998 |
| 4 | 3072.98 | 0.4500 | 0.4200 | 3088.52 | 0.4454 | 0.4246 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Wibowo, S.; Li, W.; Li, L.D. Quantitative Spectral Data Analysis Using Extreme Learning Machines Algorithm Incorporated with PCA. Algorithms 2021, 14, 18. https://0-doi-org.brum.beds.ac.uk/10.3390/a14010018
Li M, Wibowo S, Li W, Li LD. Quantitative Spectral Data Analysis Using Extreme Learning Machines Algorithm Incorporated with PCA. Algorithms. 2021; 14(1):18. https://0-doi-org.brum.beds.ac.uk/10.3390/a14010018
Chicago/Turabian StyleLi, Michael, Santoso Wibowo, Wei Li, and Lily D. Li. 2021. "Quantitative Spectral Data Analysis Using Extreme Learning Machines Algorithm Incorporated with PCA" Algorithms 14, no. 1: 18. https://0-doi-org.brum.beds.ac.uk/10.3390/a14010018





