The Interaction Between Lentiviral Integrase and LEDGF: Structural and Functional Insights
Abstract
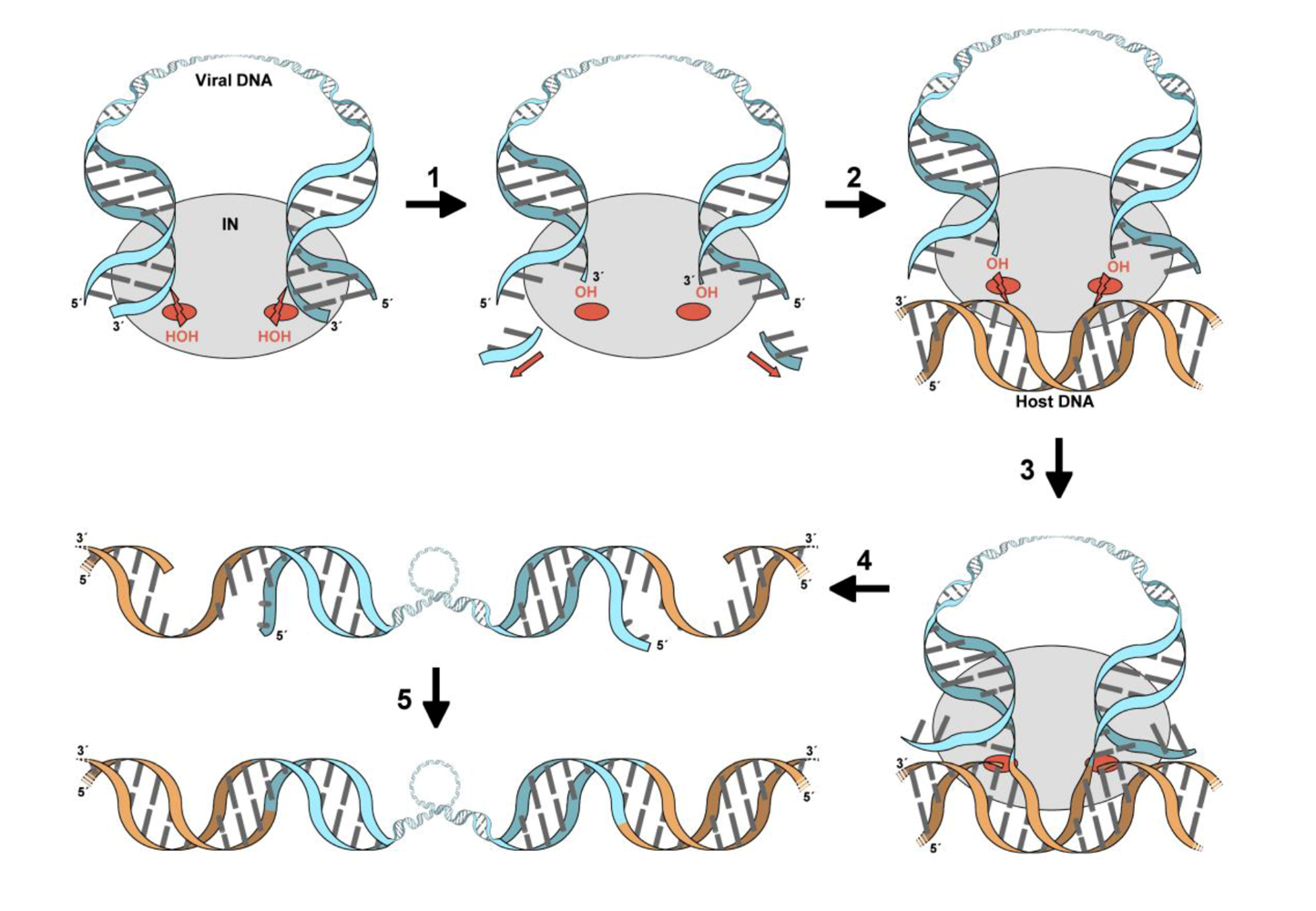
:1. Introduction
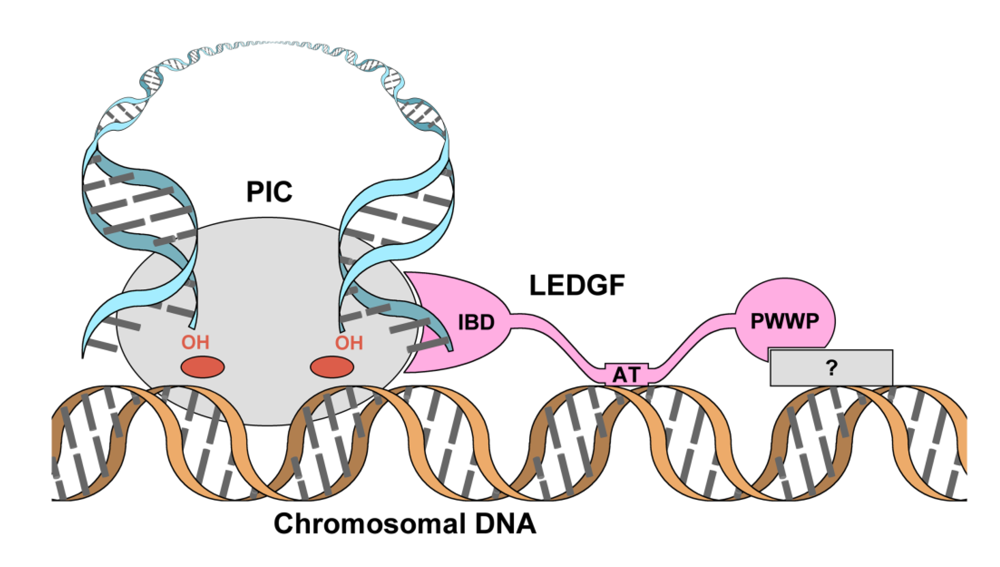
2. Domain organization of LEDGF
4. The primary IN:LEDGF interface
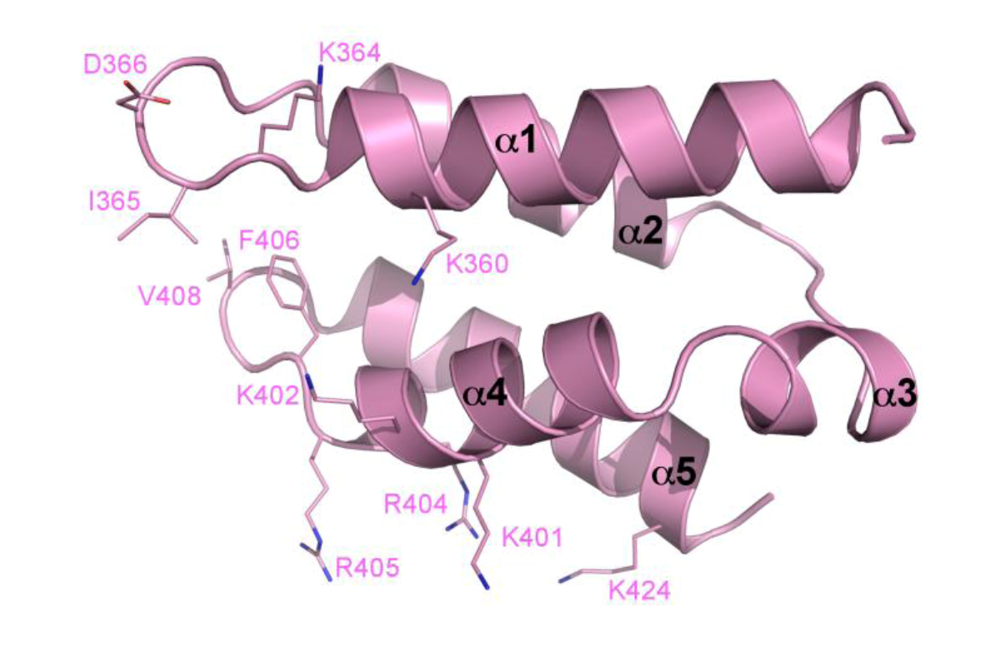
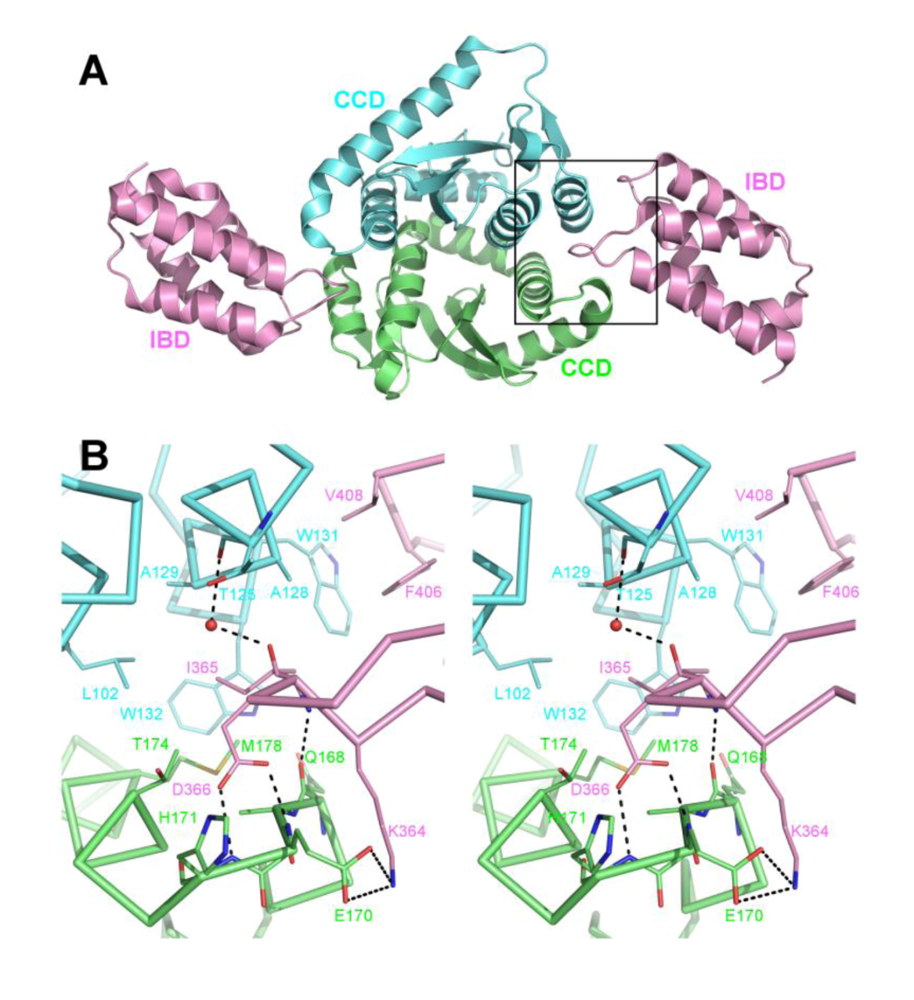
5. The IN NTD and the high affinity IN-LEDGF interaction

6. A role for LEDGF in IN tetramerization

8. Concluding remarks, remaining questions and perspectives
Acknowledgments
References and Notes
- Craigie, R. Retroviral DNA Integration. In Mobile DNA II; 2002; Craigie,, R., Gellert, M., Lambowitz, A.M., Eds.; ASM Press: Washington DC; pp. 613–630. [Google Scholar]
- Lewinski, M.K.; Bushman, F.D. Retroviral DNA integration--mechanism and consequences. Adv. Genet. 2005, 55, 147–181. [Google Scholar] [PubMed]
- Khan, E.; Mack, J.P.; Katz, R.A.; Kulkosky, J.; Skalka, A.M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991, 19, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Engelman, A.; Palmer, I.; Wingfield, P.; Craigie, R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 3428–3432. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Bushman, F.D.; Craigie, R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993, 12, 3269–3275. [Google Scholar] [PubMed]
- Engelman, A.; Craigie, R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 1992, 66, 6361–6369. [Google Scholar] [PubMed]
- Kulkosky, J.; Jones, K.S.; Katz, R.A.; Mack, J.P.; Skalka, A.M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992, 12, 2331–2338. [Google Scholar] [PubMed]
- Wlodawer, A. Crystal structures of catalytic core domains of retroviral integrases and role of divalent cations in enzymatic activity. Adv. Virus Res. 1999, 52, 335–350. [Google Scholar] [PubMed]
- Lovell, S.; Goryshin, I.Y.; Reznikoff, W.R.; Rayment, I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat. Struct. Biol. 2002, 9, 278–281. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 2005, 121, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lee, J.Y.; Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 2006, 22, 5–13. [Google Scholar] [CrossRef]
- Diamond, T.L.; Bushman, F.D. Role of metal ions in catalysis by HIV integrase analyzed using a quantitative PCR disintegration assay. Nucleic Acids Res. 2006, 34, 6116–6125. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Palmer, I.; Engelman, A.; Craigie, R.; Wingfield, P. Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J. Biol. Chem. 1994, 269, 29279–29287. [Google Scholar] [PubMed]
- Jenkins, T.M.; Engelman, A.; Ghirlando, R.; Craigie, R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996, 271, 7712–7718. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Jenkins, T.M.; Craigie, R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13659–13664. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Di Nunzio, F.; Labeja, A.; Wang, J.; Engelman, A.; Cherepanov, P. Structural basis for functional tetramerization of lentiviral integrase . PLoS Pathog. 2009, 5 , e1000515. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Hickman, A.B.; Craigie, R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994, 68, 5911–5917. [Google Scholar] [PubMed]
- van den Ent, F.M.; Vos, A.; Plasterk, R.H. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J. Virol. 1999, 73, 3176–3183. [Google Scholar] [PubMed]
- Zhao, Z.; McKee, C.J.; Kessl, J.J.; Santos, W.L.; Daigle, J.E.; Engelman, A.; Verdine, G.; Kvaratskhelia, M. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 Integrase during viral DNA binding. J. Biol. Chem. 2008, 283, 5632–5641. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.M.; Esposito, D.; Engelman, A.; Craigie, R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997, 16, 6849–6859. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Craigie, R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998, 17, 5832–5843. [Google Scholar] [CrossRef] [PubMed]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar] [PubMed]
- Eijkelenboom, A.P.; Lutzke, R.A.; Boelens, R.; Plasterk, R.H.; Kaptein, R.; Hard, K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995, 2, 807–810. [Google Scholar] [CrossRef]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997, 4, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Krucinski, J.; Miercke, L.J.; Finer-Moore, J.S.; Tang, A.H.; Leavitt, A.D.; Stroud, R.M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 8233–8238. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Ling, H.; Yang, W.; Craigie, R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001, 20, 7333–7343. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, G.; Jaskolski, M.; Alexandratos, J.; Wlodawer, A.; Merkel, G.; Katz, R.A.; Skalka, A.M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure 1996, 4, 89–96. [Google Scholar] [CrossRef]
- Yang, Z.N.; Mueser, T.C.; Bushman, F.D.; Hyde, C.C. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 2000, 296, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yan, Y.; Munshi, S.; Zugay-Murphy, J.; Xu, B.; Witmer, M.; Felock, P.; Wolfe, A.; Sardana, V.; Emini, E.A.; Hazuda, D.; Kuo, L.C. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform. J. Mol. Biol. 2000, 296, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Valkov, E.; Gupta, S.S.; Hare, S.; Helander, A.; Roversi, P.; McClure, M.; Cherepanov, P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009, 37, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Jaskolski, M.; Alexandratos, J.N.; Bujacz, G.; Wlodawer, A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009, 276, 2926–2946. [Google Scholar] [CrossRef] [PubMed]
- Bao, K.K.; Wang, H.; Miller, J.K.; Erie, D.A.; Skalka, A.M.; Wong, I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003, 278, 1323–1327. [Google Scholar] [CrossRef] [PubMed]
- Faure, A.; Calmels, C.; Desjobert, C.; Castroviejo, M.; Caumont-Sarcos, A.; Tarrago-Litvak, L.; Litvak, S.; Parissi, V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005, 33, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Mizuuchi, M.; Burke, T.R. Jr.; Craigie, R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006, 25, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Gao, K.; Bushman, F.D.; Yeager, M. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J. Mol. Biol. 2007, 366, 286–294. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.J.; Kessl, J.J.; Shkriabai, N.; Dar, M.J.; Engelman, A.; Kvaratskhelia, M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 2008, 283, 31802–31812. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Crucifix, C.; Granger, F.; Eiler, S.; Mouscadet, J.F.; Korolev, S.; Agapkina, J.; Ziganshin, R.; Gottikh, M.; Nazabal, A.; Emiliani, S.; Benarous, R.; Moras, D.; Schultz, P.; Ruff, M. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009, 28, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clercq, E.; Debyser, Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Mousnier, A.; Busschots, K.; Maroun, M.; Van Maele, B.; Tempé, D.; Vandekerckhove, L.; Moisant, F.; Ben-Slama, L.; Witvrouw, M.; Christ, F.; Rain, J.C.; Dargemont, C.; Debyser, Z.; Benarous, R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005, 280, 25517–25523. [Google Scholar] [CrossRef] [PubMed]
- Turlure, F.; Devroe, E.; Silver, P.A.; Engelman, A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004, 9, 3187–3208. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Cherepanov, P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication . PLoS Pathog. 2008, 4 , e1000046. [Google Scholar] [CrossRef] [PubMed]
- Poeschla, E.M. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 2008, 65, 1403–1424. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Devroe, E.; Silver, P.A.; Engelman, A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004, 279, 48883–48892. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.Z.; Pu, M.T.; Gowher, H.; Wu, H.P.; Ding, J.P.; Jeltsch, A.; Xu, G.L. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004, 279, 25447–25454. [Google Scholar] [CrossRef] [PubMed]
- Stec, I.; Nagl, S.B.; van Ommen, G.J.; den Dunnen, J.T. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000, 473, 1–5. [Google Scholar] [CrossRef]
- Izumoto, Y.; Kuroda, T.; Harada, H.; Kishimoto, T.; Nakamura, H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 1997, 238, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.; Cherepanov, P.; Debyser, Z.; Engelborghs, Y.; Engelman, A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 2004, 279, 33421–33429. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, M.; Llano, M.; Delgado, S.; Thompson, D.; Peretz, M.; Poeschla, E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005, 118, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Turlure, F.; Maertens, G.; Rahman, S.; Cherepanov, P.; Engelman, A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006, 34, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Vanegas, M.; Hutchins, N.; Thompson, D.; Delgado, S.; Poeschla, E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006, 360, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.; Cherepanov, P.; Pluymers, W.; Busschots, K.; De Clercq, E.; Debyser, Z.; Engelborghs, Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003, 278, 33528–33539. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.N.; Cherepanov, P.; Engelman, A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006, 119, 2563–2571. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeeusen, K.; De Rijck, J.; Busschots, K.; Desender, L.; Gijsbers, R.; Emiliani, S.; Benarous, R.; Debyser, Z.; Christ, F. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J. Mol. Biol. 2007, 372, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Cleary, M.L. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeeusen, K.; Christ, F.; Hendrix, J.; Rain, J.C.; Emiliani, S.; Benarous, R.; Debyser, Z.; Gijsbers, R.; De Rijck, J. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J. Biol. Chem. 2009, 284, 11467–11477. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Pluymers, W.; Claeys, A.; Proost, P.; De Clercq, E.; Debyser, Z. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000, 14, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Vanegas, M.; Fregoso, O.; Saenz, D.; Chung, S.; Peretz, M.; Poeschla, E.M. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004, 78, 9524–9537. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Delgado, S.; Vanegas, M.; Poeschla, E. LEDGF/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 2004, 279, 55570–55577. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.; Christ, F.; Van Maele, B.; De Rijck, J.; Gijsbers, R.; Van den Haute, C.; Witvrouw, M.; Debyser, Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006, 80, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An essential role for LEDGF/p75 in HIV integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Shun, M.C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- De Rijck, J.; Vandekerckhove, L.; Gijsbers, R.; Hombrouck, A.; Hendrix, J.; Vercammen, J.; Engelborghs, Y.; Christ, F.; Debyser, Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 2006, 80, 11498–11509. [Google Scholar] [CrossRef] [PubMed]
- Busschots, K.; Vercammen, J.; Emiliani, S.; Benarous, R.; Engelborghs, Y.; Christ, F.; Debyser, Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005, 280, 17841–17847. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007, 35, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Hematti, P.; Hong, B.K.; Ferguson, C.; Adler, R.; Hanawa, H.; Sellers, S.; Holt, I.E.; Eckfeldt, C.E.; Sharma, Y.; Schmidt, M.; von Kalle, C.; Persons, D.A.; Billings, E.M.; Verfaillie, C.M.; Nienhuis A.W.; Wolfsberg, T.G.; Dunbar, C.E.; Calmels, B. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells . PLoS Biol. 2004, 2 , e423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crise, B.; Li, Y.; Yuan, C.; Morcock, D.R.; Whitby, D.; Munroe, D.J.; Arthur, L.O.; Wu, X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005, 79, 12199–12204. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Moressi, C.J.; Scheetz, T.E.; Xie, L.; Tran, D.T.; Casavant, T.L.; Ak, P.; Benham, C.J.; Davidson, B.L.; McCray, P.B. Integration site choice of a feline immunodeficiency virus vector . J. Virol. 2006, 80 , 8820–8823. [Google Scholar] [CrossRef] [PubMed]
- Hacker, C.V.; Vink, C.A.; Wardell, T.W.; Lee, S.; Treasure, P.; Kingsman, S.M.; Mitrophanous, K.A.; Miskin, J.E. The integration profile of EIAV-based vectors. Mol Ther 2006, 14, 536–545. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, A.; Sankale, J.L.; Meloni, S.T.; Sarr, A.D.; Mboup, S.; Kanki, P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 2006, 80, 7316–7321. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.; Lewinski, M.; Ciuffi, A.; Barr, S.; Leipzig, J.; Hannenhalli, S.; Hoffmann, C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005, 3, 848–858. [Google Scholar] [CrossRef]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.M.; Ronen, K.; Berry, C.; Llano, M.; Sutherland, H.; Saenz, D.; Bickmore, W.; Poeschla, E.; Bushman, F.D. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting . PLoS One 2007, 2 , e1340. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, M.K.; Bisgrove, D.; Shinn, P.; Chen, H.; Hoffmann, C.; Hannenhalli, S.; Verdin, E.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005, 79, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; Sorensen, R.; Forster, A.; Fraser, P.; Cohen, J.I.; de Saint Basile, G.; Alexander, I.; Wintergerst, U.; Frebourg, T.; Aurias, A.; Stoppa-Lyonnet, D.; Romana, S.; Radford-Weiss, I.; Gross, F.; Valensi, F.; Delabesse, E.; Macintyre, E.; Sigaux, F. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Howe, S.J.; Mansour, M.R.; Schwarzwaelder, K.; Bartholomae, C.; Hubank, M.; Kempski, H.; Brugman, M.H.; Pike-Overzet, K.; Chatters, S.J.; de Ridder, D.; Gilmour, K.C.; Adams, S.; Thornhill, S.I.; Parsley, K.L.; Staal, F.J.; Gale, R.E.; Linch, D.C.; Bayford, J.; Brown, L.; Quaye, M.; Kinnon, C.; Ancliff, P.; Webb, D.K.; Schmidt, M.; von Kalle, C.; Gaspar, H.B.; Thrasher, A.J. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008, 118, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Diamond, T.L.; Hwang, Y.; Marshall, H.M.; Bushman, F.D. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 2006, 17, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Meehan, A.M.; Saenz, D.T.; Morrison, J.H.; Garcia-Rivera, J.A.; Peretz, M.; Llano, M.; Poeschla, E.M. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors . PLoS Pathog. 200, 5 , e1000522. [Google Scholar] [CrossRef]
- Cherepanov, P.; Sun, Z.Y.; Rahman, S.; Maertens, G.; Wagner, G.; Engelman, A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005, 12, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Ambrosio, A.L.; Rahman, S.; Ellenberger, T.; Engelman, A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 17308–17313. [Google Scholar] [CrossRef] [PubMed]
- Busschots, K.; Voet, A.; De Maeyer, M.; Rain, J.C.; Emiliani, S.; Benarous, R.; Desender, L.; Debyser, Z.; Christ, F. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J. Mol. Biol. 2007, 365, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Lu, R.; Vandegraaff, N.; Cherepanov, P.; Engelman, A. Structure-based mutagenesis of the integrase-LEDGF/p75 interface uncouples a strict correlation between in vitro protein binding and HIV-1 fitness. Virology 2007, 357, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Bouyac-Bertoia, M.; Dvorin, J.D.; Fouchier, R.A.; Jenkins, Y.; Meyer, B.E.; Wu, L.I.; Emerman, M.; Malim, M.H. HIV-1 infection requires a functional integrase NLS. Mol. Cell 2001, 7, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Limon, A.; Devroe, E.; Lu, R.; Ghory, H.Z.; Silver, P.A.; Engelman, A. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 2002, 76, 10598–10607. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Limon, A.; Devroe, E.; Silver, P.A.; Cherepanov, P.; Engelman, A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 2004, 78, 12735–12746. [Google Scholar] [CrossRef] [PubMed]
- Molteni, V.; Greenwald, J.; Rhodes, D.; Hwang, Y.; Kwiatkowski, W.; Bushman, F.D.; Siegel, J.S.; Choe, S. Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr. D. Biol. Crystallogr. 2001, 57, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, Y.; Chen, J.; Yang, L.; Zheng, Y.; Tang, Y.; Shen, X.; Jiang, H. D77, one benzoic acid derivative, functions as a novel anti-HIV-1 inhibitor targeting the interaction between integrase and cellular LEDGF/p75. Biochem. Biophys. Res. Commun. 2008, 375, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; McGuinness, D.E.; Prongay, A.J.; Feld, B.; Ingravallo, P.; Ogert, R.A.; Lunn, C.A.; Howe, J.A. Screening for antiviral inhibitors of the HIV integrase-LEDGF/p75 interaction using the AlphaScreen luminescent proximity assay. J Biomol Screen 2008, 13, 406–414. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Barreca, M.L.; Ferro, S.; Christ, F.; Iraci, N.; Gitto, R.; Monforte, A.M.; Debyser, Z.; Chimirri, A. Pharmacophore-based discovery of small-molecule inhibitors of protein-protein interactions between HIV-1 integrase and cellular cofactor LEDGF/p75. ChemMedChem 2009, 4, 1311–1316. [Google Scholar] [CrossRef] [Green Version]
- Hare, S.; Shun, M.C.; Gupta, S.S.; Valkov, E.; Engelman, A.; Cherepanov, P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75 . PLoS Pathog. 2009, 5 , e1000259. [Google Scholar] [CrossRef] [PubMed]
- Maignan, S.; Guilloteau, J.P.; Zhou-Liu, Q.; Clement-Mella, C.; Mikol, V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 1998, 282, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Berthoux, L.; Sebastian, S.; Muesing, M.A.; Luban, J. The role of lysine 186 in HIV-1 integrase multimerization. Virology 2007, 364, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Hayouka, Z.; Rosenbluh, J.; Levin, A.; Loya, S.; Lebendiker, M.; Veprintsev, D.; Kotler, M.; Hizi, A.; Loyter, A.; Friedler, A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 8316–8321. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Craigie, R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 2005, 280, 29334–29339. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Pursley, M.H.; Grandgenett, D.P. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002, 76, 3105–3113. [Google Scholar] [CrossRef] [PubMed]
- Bosserman, M.A.; O'Quinn, D.F.; Wong, I. Loop202-208 in avian sarcoma virus integrase mediates tetramer assembly and processing activity. Biochemistry 2007, 46, 11231–11239. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Jones, G.S.; Hung, M.; Wagner, A.H.; MacArthur, H.L.; Liu, X.; Leavitt, S.; McDermott, M.J.; Tsiang, M. HIV-1 integrase preassembled on donor DNA is refractory to activity stimulation by LEDGF/p75. Biochemistry 2007, 46, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.K.; Sinha, S.; Grandgenett, D.P. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 2007, 81, 3969–3979. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, N.K.; Engelman, A. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 2007, 360, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Botbol, Y.; Raghavendra, N.K.; Rahman, S.; Engelman, A.; Lavigne, M. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008, 36, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Bushman, F.D. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006, 22, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Maddali, K.; Metifiot, M.; Pommier, Y. HIV-1 IN Inhibitors: 2010 Update and Perspectives . Curr Top Med Chem 2009, . [Google Scholar]
- Walker, M.A. New approaches for inhibiting HIV integrase: a journey beyond the active site. Curr Opin Investig Drugs 2009, 10, 129–136. [Google Scholar] [PubMed]
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Zhou, Y.; Elleder, D.; Diamond, T.L.; Bonamy, G.M.; Irelan, J.T.; Chiang, C.Y.; Tu, B.P.; De Jesus, P.D.; Lilley, C.E.; Seidel, S.; Opaluch, A.M.; Caldwell, J.S.; Weitzman, M.D.; Kuhen, K.L.; Bandyopadhyay, S.; Ideker, T.; Orth, A.P.; Miraglia, L.J.; Bushman, F.D.; Young, J.A.; Chanda, S.K. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, M.; Huang, Q.; Gates, A.T.; Zhang, X.D.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.J.; Espeseth, A.S. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 2008, 4, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D.; Malani, N.; Fernandes, J.; D'Orso, I.; Cagney, G.; Diamond, T.L.; Zhou, H.; Hazuda, D.J.; Espeseth, A.S.; König, R.; Bandyopadhyay, S.; Ideker, T.; Goff, S.P.; Krogan, N.J.; Frankel, A.D.; Young, J.A.; Chanda, S.K. Host cell factors in HIV replication: meta-analysis of genome-wide studies . PLoS Pathog. 2009, 5 , e1000437. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.L.; Houzet, L.; Yedavalli, V.S.; Jeang, K.T. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 2009, 284, 19463–19473. [Google Scholar] [CrossRef] [PubMed]
- Christ, F.; Thys, W.; De Rijck, J.; Gijsbers, R.; Albanese, A.; Arosio, D.; Emiliani, S.; Rain, J.C.; Benarous, R.; Cereseto, A.; Debyser, Z. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 2008, 18, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Studamire, B.; Goff, S.P. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 2008, 5, 48. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Hare, S.; Cherepanov, P. The Interaction Between Lentiviral Integrase and LEDGF: Structural and Functional Insights. Viruses 2009, 1, 780-801. https://0-doi-org.brum.beds.ac.uk/10.3390/v1030780
Hare S, Cherepanov P. The Interaction Between Lentiviral Integrase and LEDGF: Structural and Functional Insights. Viruses. 2009; 1(3):780-801. https://0-doi-org.brum.beds.ac.uk/10.3390/v1030780
Chicago/Turabian StyleHare, Stephen, and Peter Cherepanov. 2009. "The Interaction Between Lentiviral Integrase and LEDGF: Structural and Functional Insights" Viruses 1, no. 3: 780-801. https://0-doi-org.brum.beds.ac.uk/10.3390/v1030780