Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses
Abstract
:1. Introduction
2. Methods
2.1. Dataset
2.2. Multiple Sequence Alignments and Phylogenetic Analysis
2.3. Detection of Chimeric Rep Proteins
2.4. Sequence Logos
3. Results and Discussion
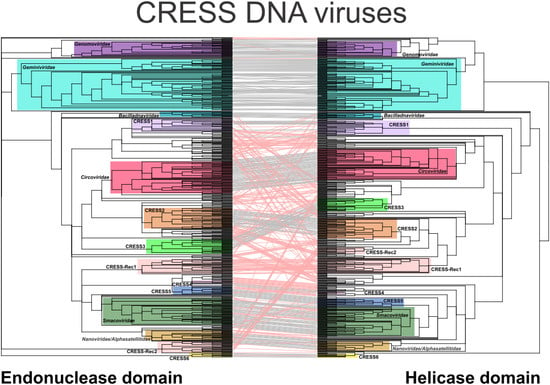
3.1. Evaluation of Coevolution of the Nuclease and Helicase Domains
3.2. Coherent Evolutionary Patterns in Classified CRESS DNA Viruses
3.3. High Incidence of Chimerism in the Reps of Unclassified CRESS DNA Viruses
3.4. Six Potentially New Families of CRESS DNA Viruses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dennehy, J.J. Evolutionary ecology of virus emergence. Ann. N. Y. Acad. Sci. 2017, 1389, 124–146. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Drummond, A.J. The evolutionary genetics of viral emergence. Curr. Top. Microbiol. Immunol. 2007, 315, 51–66. [Google Scholar] [PubMed]
- Lefeuvre, P.; Moriones, E. Recombination as a motor of host switches and virus emergence: Geminiviruses as case studies. Curr. Opin. Virol. 2015, 10, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef] [PubMed]
- Iranzo, J.; Krupovic, M.; Koonin, E.V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 2016, 7, e00978-16. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 2013, 3, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; de la Cruz, F.; Dyda, F.; Hickman, A.B.; Moncalian, G.; Ton-Hoang, B. Breaking and joining single-stranded DNA: The HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013, 11, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Koonin, E.V.; Wolf, Y.I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990, 262, 145–148. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Dayaram, A.; Kraberger, S.; Goldstien, S.; Varsani, A.; Krupovic, M. Evolutionary history of ssDNA bacilladnaviruses features horizontal acquisition of the capsid gene from ssRNA nodaviruses. Virology 2017, 504, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Breitbart, M. Exploring the viral world through metagenomics. Curr. Opin. Virol. 2011, 1, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benko, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Holmes, E.C. Validation of high rates of nucleotide substitution in geminiviruses: Phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 2009, 90 Pt 6, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef] [PubMed]
- Grigoras, I.; Timchenko, T.; Grande-Perez, A.; Katul, L.; Vetten, H.J.; Gronenborn, B. High variability and rapid evolution of a nanovirus. J. Virol. 2010, 84, 9105–9117. [Google Scholar] [CrossRef] [PubMed]
- Harkins, G.W.; Delport, W.; Duffy, S.; Wood, N.; Monjane, A.L.; Owor, B.E.; Donaldson, L.; Saumtally, S.; Triton, G.; Briddon, R.W.; et al. Experimental evidence indicating that mastreviruses probably did not co-diverge with their hosts. Virol. J. 2009, 6, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefeuvre, P.; Lett, J.M.; Varsani, A.; Martin, D.P. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 2009, 83, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Biagini, P.; Lefeuvre, P.; Golden, M.; Roumagnac, P.; Varsani, A. Recombination in eukaryotic single stranded DNA viruses. Viruses 2011, 3, 1699–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Koonin, E.V. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci. Rep. 2014, 4, 5347. [Google Scholar] [CrossRef] [PubMed]
- Stedman, K.M. Deep recombination: RNA and ssDNA virus genes in DNA virus and host genomes. Annu. Rev. Virol. 2015, 2, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Diemer, G.S.; Stedman, K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct 2012, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Bronner, G.; Vaulot, D.; Forterre, P.; Krupovic, M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 2013, 4, 2700. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Zhi, N.; Li, J.; Hu, G.; Koonin, E.V.; Wong, S.; Shevchenko, S.; Zhao, K.; Young, N.S. Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol. Evol. 2015, 7, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Quaiser, A.; Krupovic, M.; Dufresne, A.; Francez, A.J.; Roux, S. Diversity and comparative genomics of chimeric viruses in Sphagnum-dominated peatlands. Virus Evol. 2016, 2, vew025. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Krupovic, M. Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol. 2017, 3, vew037. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Krupovic, M. Smacoviridae: A new family of animal-associated single-stranded DNA viruses. Arch. Virol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Lefeuvre, P.; Varsani, A.; Hoareau, M.; Semegni, J.Y.; Dijoux, B.; Vincent, C.; Reynaud, B.; Lett, J.M. Complex recombination patterns arising during geminivirus coinfections preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 2011, 7, e1002203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monjane, A.L.; Martin, D.P.; Lakay, F.; Muhire, B.M.; Pande, D.; Varsani, A.; Harkins, G.; Shepherd, D.N.; Rybicki, E.P. Extensive recombination-induced disruption of genetic interactions is highly deleterious but can be partially reversed by small numbers of secondary recombination events. J. Virol. 2014, 88, 7843–7851. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Golden, M.; Murrell, B.; Lefeuvre, P.; Lett, J.M.; Gray, A.; Poon, A.Y.; Ngandu, N.K.; Semegni, Y.; Tanov, E.P.; et al. Evidence of pervasive biologically functional secondary structures within the genomes of eukaryotic single-stranded DNA viruses. J. Virol. 2014, 88, 1972–1989. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Martin, D.P.; Roumagnac, P.; Navas-Castillo, J.; Fiallo-Olivé, E.; Moriones, E.; Lett, J.-M.; Zerbini, F.M.; Varsani, A. Alphasatellitidae: A new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch. Virol. 2018, in press. [Google Scholar]
- Yoshida, M.; Mochizuki, T.; Urayama, S.I.; Yoshida-Takashima, Y.; Nishi, S.; Hirai, M.; Nomaki, H.; Takaki, Y.; Nunoura, T.; Takai, K. Quantitative Viral Community DNA Analysis Reveals the Dominance of Single-Stranded DNA Viruses in Offshore Upper Bathyal Sediment from Tohoku, Japan. Front. Microbiol. 2018, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Goldstien, S.; Arguello-Astorga, G.R.; Zawar-Reza, P.; Gomez, C.; Harding, J.S.; Varsani, A. Diverse small circular DNA viruses circulating amongst estuarine molluscs. Infect. Genet. Evol. 2015, 31, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Ng, T.F.; Lager, K.M.; Alt, D.P.; Delwart, E.; Pogranichniy, R.M. Identification of several clades of novel single-stranded circular DNA viruses with conserved stem-loop structures in pig feces. Arch. Virol. 2015, 160, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Zhang, W.; Sachsenroder, J.; Kondov, N.O.; da Costa, A.C.; Vega, E.; Holtz, L.R.; Wu, G.; Wang, D.; Stine, C.O.; et al. A diverse group of small circular ssDNA viral genomes in human and non-human primate stools. Virus Evol. 2015, 1, vev017. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; da Costa, A.C.; Del Valle Mendoza, J.; Bucardo-Rivera, F.; Nordgren, J.; O’Ryan, M.; Deng, X.; Delwart, E. The fecal virome of South and Central American children with diarrhea includes small circular DNA viral genomes of unknown origin. Arch. Virol. 2016, 161, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Mahmood, A.; Yang, S.; Wang, X.; Shen, Q.; Shan, T.; Deng, X.; Li, J.; Hua, X.; et al. Plasma virome of cattle from forest region revealed diverse small circular ssDNA viral genomes. Virol. J. 2018, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Dayaram, A.; Marinov, M.; Ware, J.; Kraberger, S.; Stainton, D.; Breitbart, M.; Varsani, A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J. Gen. Virol. 2012, 93 Pt 12, 2668–2681. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Duffy, S.; Breitbart, M. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 2009, 90 Pt 10, 2418–2424. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Schenck, R.O.; Harbeitner, R.C.; Lawler, S.N.; Breitbart, M. Novel circular single-stranded DNA viruses identified in marine invertebrates reveal high sequence diversity and consistent predicted intrinsic disorder patterns within putative structural proteins. Front. Microbiol. 2015, 6, 696. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Galatowitsch, M.L.; Arguello-Astorga, G.R.; van Bysterveldt, K.; Kraberger, S.; Stainton, D.; Harding, J.S.; Roumagnac, P.; Martin, D.P.; Lefeuvre, P.; et al. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect. Genet. Evol. 2016, 39, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Blinkova, O.; Victoria, J.; Li, Y.; Keele, B.F.; Sanz, C.; Ndjango, J.B.; Peeters, M.; Travis, D.; Lonsdorf, E.V.; Wilson, M.L.; et al. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 2010, 91 Pt 1, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Ghabrial, S.A.; Jiang, D.; Varsani, A. Genomoviridae: A new family of widespread single-stranded DNA viruses. Arch. Virol. 2016, 161, 2633–2643. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazlauskas, D.; Varsani, A.; Krupovic, M. Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses. Viruses 2018, 10, 187. https://0-doi-org.brum.beds.ac.uk/10.3390/v10040187
Kazlauskas D, Varsani A, Krupovic M. Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses. Viruses. 2018; 10(4):187. https://0-doi-org.brum.beds.ac.uk/10.3390/v10040187
Chicago/Turabian StyleKazlauskas, Darius, Arvind Varsani, and Mart Krupovic. 2018. "Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses" Viruses 10, no. 4: 187. https://0-doi-org.brum.beds.ac.uk/10.3390/v10040187