The Effect of a Commercially Available Bacteriophage and Bacteriocin on Listeria monocytogenes in Coleslaw
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteriophage Propagation and Bacteriophage Assays
2.2. Host Range of P100
2.3. Coleslaw Food Trial with P100
2.4. Rate of Resistance to P100
2.5. P100 and Nisaplin® Checkerboard Assay in Broth
2.6. P100 and Nisaplin® Checkerboard Assay in Coleslaw Liquid
2.7. Coleslaw Food Trial with P100 and Nisaplin® in Combination
2.8. Statistical Analysis
3. Results
3.1. Host Range of P100
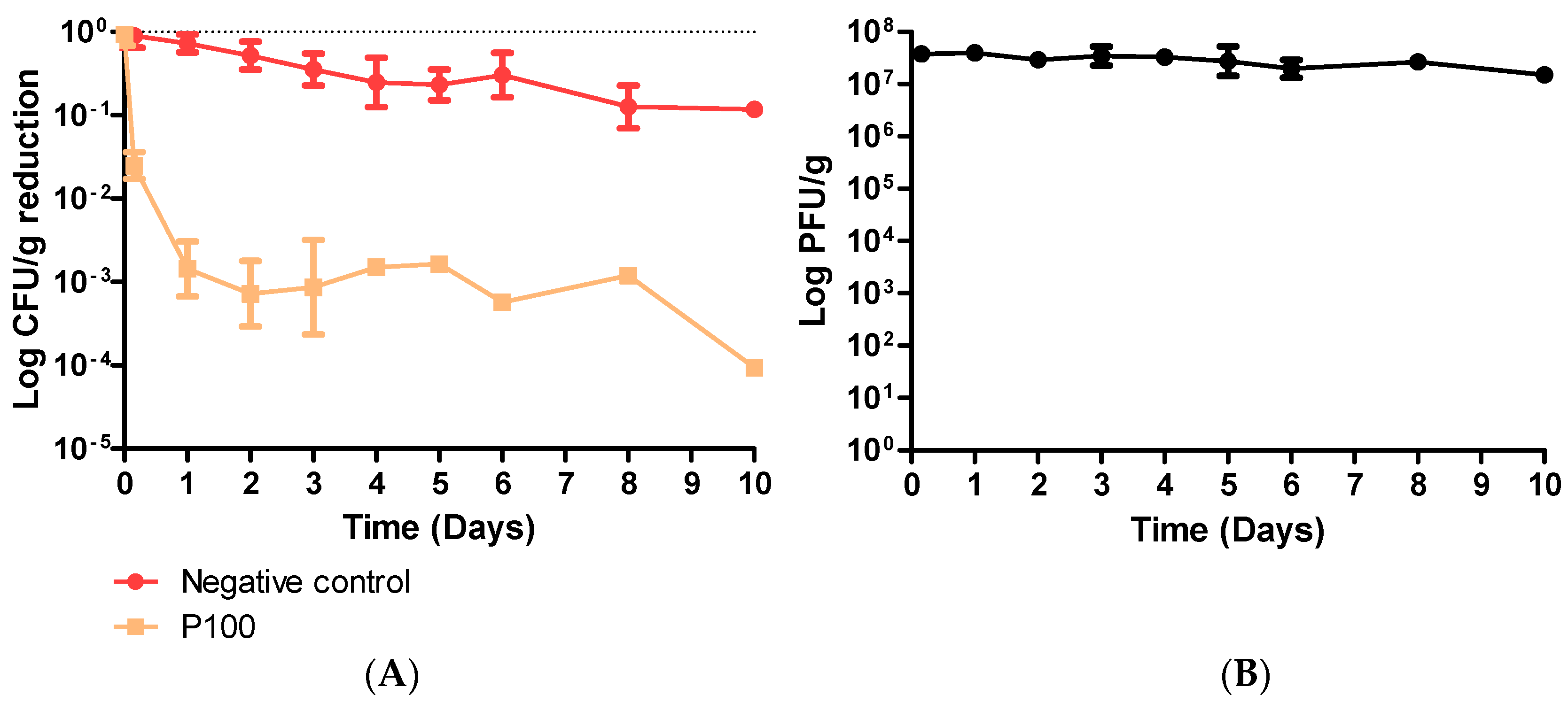
3.2. Coleslaw Food Trial with P100
3.3. P100 and Nisaplin® Checkerboard Assays in Broth and Coleslaw
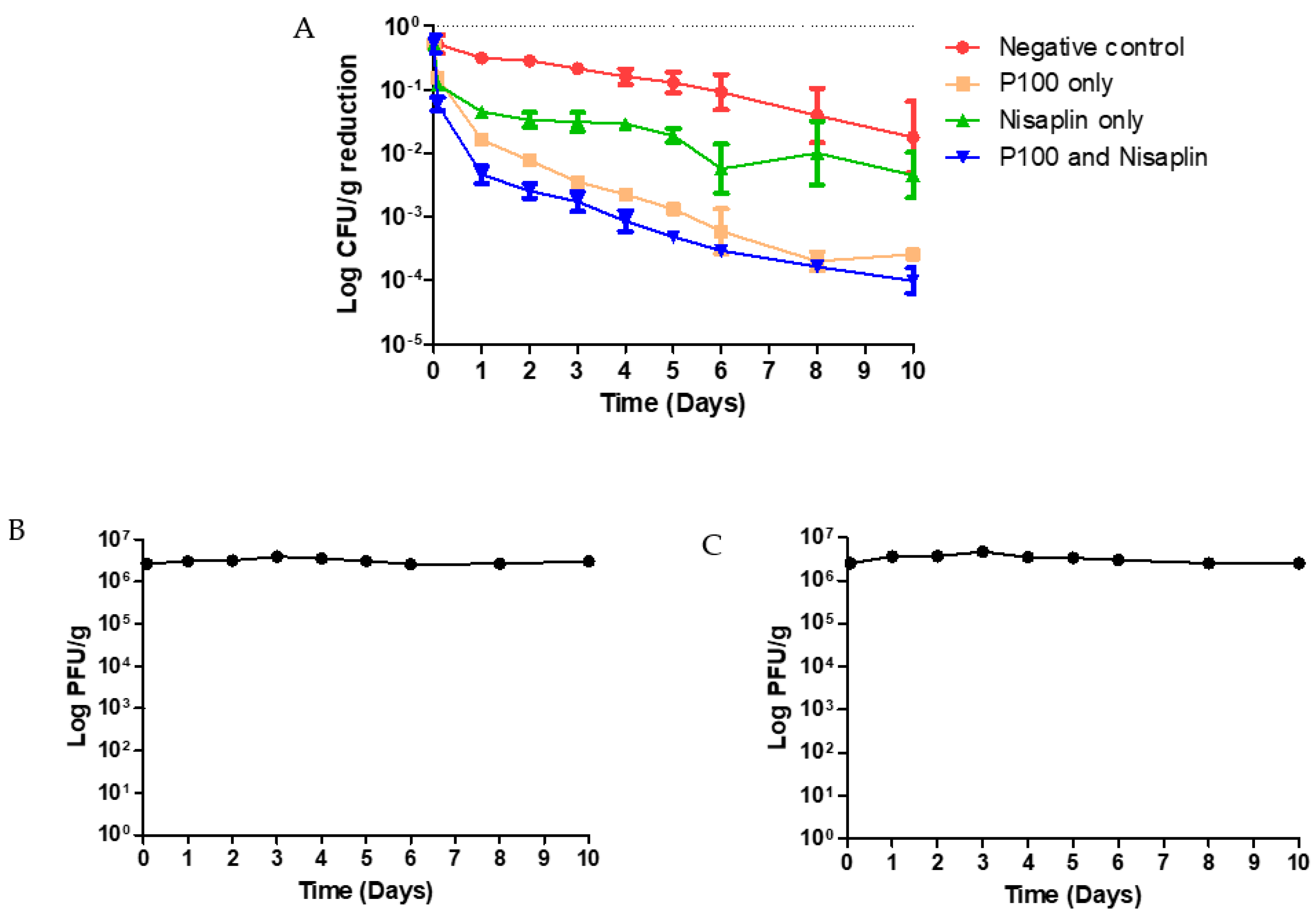
3.4. Coleslaw Food Trial with P100 and Nisaplin® in Combination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hernandez-Milian, A.; Payeras-Cifre, A. What is new in listeriosis? Biomed. Res. Int. 2014, 2014, 358051. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Nørrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the eu. EFSA J. 2018, 16, e05134. [Google Scholar]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Todd, E.C.D.; Notermans, S. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control. 2011, 22, 1484–1490. [Google Scholar] [CrossRef]
- García, P.; Rodríguez, L.; Rodríguez, A.; Martínez, B. Food biopreservation: Promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci. Technol. 2010, 21, 373–382. [Google Scholar] [CrossRef]
- Asioli, D.; Aschemann-Witzel, J.; Caputo, V.; Vecchio, R.; Annunziata, A.; Næs, T.; Varela, P. Making sense of the “clean label” trends: A review of consumer food choice behavior and discussion of industry implications. Food Res. Int. 2017, 99, 58–71. [Google Scholar] [CrossRef]
- Mahony, J.; Mauliffe, O.; Ross, R.P.; van Sinderen, D. Bacteriophages as biocontrol agents of food pathogens. Curr. Opin. Biotechnol. 2011, 22, 157–163. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777. [Google Scholar] [CrossRef]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.; de Haas, E.; Neve, H.; Strain, R.; Cousin, F.J.; Stockdale, S.R.; Ross, R.P.; Hill, C. Isolation of a novel phage with activity against streptococcus mutans biofilms. PloS ONE 2015, 10, e0138651. [Google Scholar] [CrossRef] [PubMed]
- Draper, L.A.; Cotter, P.D.; Hill, C.; Ross, R.P. The two peptide lantibiotic lacticin 3147 acts synergistically with polymyxin to inhibit gram negative bacteria. BMC Microbiol. 2013, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Carlton, R.M.; Noordman, W.H.; Biswas, B.; de Meester, E.D.; Loessner, M.J. Bacteriophage p100 for control of listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 2005, 43, 301–312. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Bhunia, A.K. Adhesion, invasion, and translocation characteristics of listeria monocytogenes serotypes in caco-2 cell and mouse models. Appl. Environ. Microbiol. 2003, 69, 3640–3645. [Google Scholar] [CrossRef]
- Clayton, E.M.; Hill, C.; Cotter, P.D.; Ross, R.P. Real-time pcr assay to differentiate listeriolysin s-positive and -negative strains of listeria monocytogenes. Appl. Environ. Microbiol. 2011, 77, 163–171. [Google Scholar] [CrossRef]
- Fox, E.M.; Leonard, N.; Jordan, K. Physiological and transcriptional characterization of persistent and nonpersistent listeria monocytogenes isolates. Appl. Environ. Microbiol. 2011, 77, 6559–6569. [Google Scholar] [CrossRef]
- Murray, E.G.D.; Webb, R.A.; Swann, M.B.R. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n.Sp.). J. Pathol. Bacteriol. 1926, 29, 407–439. [Google Scholar] [CrossRef]
- Lianou, A.; Sofos, J.N. A review of the incidence and transmission of Listeria monocytogenes in ready-to-eat products in retail and food service environments. J. Food Prot. 2007, 70, 2172–2198. [Google Scholar] [CrossRef]
- Leistner, L.; Gorris, L.G.M. Food preservation by hurdle technology. Trends Food Sci. Technol. 1995, 6, 41–46. [Google Scholar] [CrossRef]
- Wu, F.M.; Beuchat, L.R.; Doyle, M.P.; Garrett, V.; Wells, J.G.; Swaminathan, B. Fate of escherichia coli o157:H7 in coleslaw during storage. J. Food Prot. 2002, 65, 845–847. [Google Scholar] [CrossRef] [PubMed]
- George, A.E.; Levett, P.N. Effect of temperature and ph on survival of Listeria monocytogenes in coleslaw. Int. J. Food Microbiol 1990, 11, 345–349. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Weber, S.; Hammes, W.P. Effect of ecological factors on the inhibitory spectrum and activity of bacteriocins. Int. J. Food Microbiol. 1999, 46, 207–217. [Google Scholar] [CrossRef]
- Oliveira, M.; Viñas, I.; Colàs, P.; Anguera, M.; Usall, J.; Abadias, M. Effectiveness of a bacteriophage in reducing listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiol. 2014, 38, 137–142. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards. Evaluation of the safety and efficacy of listex™ p100 for reduction of pathogens on different ready-to-eat (rte) food products. EFSA J. 2016, 14, e04565. [Google Scholar]
- Hagens, S.; Loessner, M.J. Bacteriophage for biocontrol of foodborne pathogens: Calculations and considerations. Curr. Pharm. Biotechnol. 2010, 11, 58–68. [Google Scholar] [CrossRef]
- Soni, K.A.; Nannapaneni, R.; Hagens, S. Reduction of listeria monocytogenes on the surface of fresh channel catfish fillets by bacteriophage listex p100. Foodborne Pathog. Dis. 2010, 7, 4. [Google Scholar] [CrossRef]
- Migueis, S.; Saraiva, C.; Esteves, A. Efficacy of listex p100 at different concentrations for reduction of listeria monocytogenes inoculated in sashimi. J. Food Prot. 2017, 80, 2094–2098. [Google Scholar] [CrossRef]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef]
- Holck, A.; Berg, J. Inhibition of listeria monocytogenes in cooked ham by virulent bacteriophages and protective cultures. Appl Env. Microbiol 2009, 75, 6944–6946. [Google Scholar] [CrossRef]
- Leverentz, B.; Conway, W.S.; Camp, M.J.; Janisiewicz, W.J.; Abuladze, T.; Yang, M.; Saftner, R.; Sulakvelidze, A. Biocontrol of listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 2003, 69, 4519–4526. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Almeida, R. Antibacterial efficacy of nisin, bacteriophage p100 and sodium lactate against Listeria monocytogenes in ready-to-eat sliced pork ham. Braz. J. Microbiol 2017, 48, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Tokman, J.I.; Kent, D.J.; Wiedmann, M.; Denes, T. Temperature significantly affects the plaquing and adsorption efficiencies of listeria phages. Front. Microbiol. 2016, 7, 631. [Google Scholar] [CrossRef] [PubMed]
- Hagens, S.; Loessner, M.J. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 2007, 76, 513–519. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli o157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Chaturongakul, S.; Ounjai, P. Phage–host interplay: Examples from tailed phages and gram-negative bacterial pathogens. Front. Microbiol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Abuladze, T.; Li, M.; Menetrez, M.Y.; Dean, T.; Senecal, A.; Sulakvelidze, A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by escherichia coli o157:H7. Appl. Environ. Microbiol. 2008, 74, 6230–6238. [Google Scholar] [CrossRef]
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Chapter six—Bacteriophages for detection and control of bacterial pathogens in food and food-processing environment. In Advances in Food and Nutrition Research; Henry, J., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 67, pp. 241–288. [Google Scholar]
- Guenther, S.; Loessner, M.J. Bacteriophage biocontrol of Listeria monocytogenes on soft ripened white mold and red-smear cheeses. Bacteriophage 2011, 1, 94–100. [Google Scholar] [CrossRef]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Diez-Gonzalez, F. Reduction of Escherichia coli o157:H7 viability on leafy green vegetables by treatment with a bacteriophage mixture and trans-cinnamaldehyde. Food Microbiol. 2011, 28, 149–157. [Google Scholar] [CrossRef]
- Harris, L.J.; Fleming, H.P.; Klaenhammer, T.R. Sensitivity and resistance of listeria monocytogenes atcc 19115, scott a, and ual500 to nisin. J. Food Prot. 1991, 54, 836–840. [Google Scholar] [CrossRef] [PubMed]
| Strain | Equivalent Names | Origin | Serotype | Efficiency of Plaquing of P100 |
|---|---|---|---|---|
| L. monocytogenes ATCC 35152 | Guinea pig [15] | 1/2a | 1.22 ± 0.4 | |
| L. monocytogenes 33116 | ATCC 19117 | Animal [16] | 4d | 1 ± 0 |
| L. monocytogenes 33120 | ATCC 19118 | Animal [16] | 4e | 0.92 ± 0.54 |
| L. innocua DPC 3372 | - | 0.83 ± 0.25 | ||
| L. monocytogenes 33176 | 20240-954 | Animal [16] | 1/2b | 0.72 ± 0.31 |
| L. monocytogenes 33104 | F2365, JI-119, TS43 | California outbreak, 1985 [16] | 4b | 0.70 ± 0.03 |
| L. monocytogenes ScottA | 33013 | Clinical (Massachusetts outbreak, 1983) [16] | 4b | 0.70 ± 0.03 |
| L. monocytogenes EGDE | Rabbit [17] | 1/2a | 0.61 ± 0.2 | |
| L. monocytogenes 33007 | RM2218 | Food [16] | 4b | 0.59 ± 0.22 |
| L. monocytogenes 33186 | 20674-01 | Animal [16] | 1/2b | 0.49 ± 0.25 |
| L. monocytogenes ATCC 15313 | Rabbit [18] | 1/2c | 0.29 ± 0.02 | |
| L. monocytogenes ATCC 19112 | Human CSF [15] | 1/2c | 0.24 ± 0.06 | |
| L. monocytogenes 6179 | Cheese; production environment [17] | 1/2a | 0.002 ± 0.001 | |
| L. grayi CD671 | ATCC 25400 | Corn stalks [16] | - | - |
| L. monocytogenes 33028 | OB001102 | Food [16] | 1/2b | - |
| L. innocua FA2039 | - | - |
| Antimicrobial Concentration | P100 | P100 | P100 | P100 | P100 |
|---|---|---|---|---|---|
| MOI 0 | MOI 0.01 | MOI 0.1 | MOI 1 | MOI 10 | |
| Nisaplin® 0 µg/mL | 9.6 × 105 | 1.2 × 106 | 1.1 × 106 | 1.3 × 105 | <1.0 × 104 MIC P100 Alone |
| Nisaplin® 12.5 µg/mL | 5.0 × 105 | 4.6 × 105 | 6.8 × 105 | 6.2 × 104 | <1.0 × 104 FIC 1.25 Indifferent |
| Nisaplin® 25 µg/mL | 4.2 × 104 | 8.0 × 104 | 2.0 × 104 | <1.0 × 104 FIC 0.6 Partial synergy | <1.0 × 104 FIC 1.5 Indifferent |
| Nisaplin® 50 µg/mL | <1.0 × 104 MIC Nisaplin® Alone | <1.0 × 104 FIC 1.001 Indifferent | <1.0 × 104 FIC 1.01 Indifferent | <1.0 × 104 FIC 1.1 Indifferent | <1.0 × 104 FIC 2 Indifferent |
| Nisaplin® 100 µg/mL | <1.0 × 104 FIC 2 Indifferent | <1.0 × 104 FIC 2.001 Indifferent | <1.0 × 104 FIC 2.01 Indifferent | <1.0 × 104 FIC 2.1 Indifferent | <1.0 × 104 FIC 3 Indifferent |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewis, R.; Bolocan, A.S.; Draper, L.A.; Ross, R.P.; Hill, C. The Effect of a Commercially Available Bacteriophage and Bacteriocin on Listeria monocytogenes in Coleslaw. Viruses 2019, 11, 977. https://0-doi-org.brum.beds.ac.uk/10.3390/v11110977
Lewis R, Bolocan AS, Draper LA, Ross RP, Hill C. The Effect of a Commercially Available Bacteriophage and Bacteriocin on Listeria monocytogenes in Coleslaw. Viruses. 2019; 11(11):977. https://0-doi-org.brum.beds.ac.uk/10.3390/v11110977
Chicago/Turabian StyleLewis, Rhea, Andrei Sorin Bolocan, Lorraine A. Draper, R. Paul Ross, and Colin Hill. 2019. "The Effect of a Commercially Available Bacteriophage and Bacteriocin on Listeria monocytogenes in Coleslaw" Viruses 11, no. 11: 977. https://0-doi-org.brum.beds.ac.uk/10.3390/v11110977





