Characterization of the Role of Host Cellular Factor Histone Deacetylase 10 during HIV-1 Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid, Chemicals, and Antibodies
2.2. Cell Culture and Primary Cell Isolation
2.3. Generation of Virus and Infection
2.4. Establishment of HDAC10 Knockdown Cell Line
2.5. Water-Soluble Tetrazolium Salt (WST-1) Assay
2.6. Measurement of Transcription by Real-Time Quantitative RT-PCR
2.7. Measurement of Total Viral DNA, Integrated DNA, and 2-LTR Viral DNA by Real-Time Quantitative PCR
2.8. Luciferase Assay and Western Blot (WB)
2.9. Co-Immunoprecipitation (Co-IP) Assay
2.10. Statistical Analysis
3. Results
3.1. Endogenous HDAC10 Is Downregulated During HIV-1 Replication
3.2. HDAC10 Downregulation Benefits Viral Replication
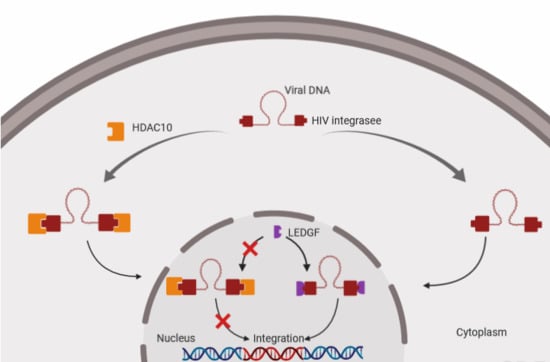
3.3. The Downregulation of HDAC10 Facilitates Viral Integration
3.4. HDAC10 Interacts with HIV-1 IN Through Binding to the Region of Amino Acids 55 to 165 of IN
3.5. The Presence of HDAC10 Weakens the Interaction of HIV-1 IN with LEDGF/p75
3.6. The Downregulation of Endogenous HDAC10 Enhances the Infectiousness of Progeny Virus
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Herbein, G.; Wendling, D. Histone deacetylases in viral infections. Clin. Epigenetics 2010, 1, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirakawa, K.; Chavez, L.; Hakre, S.; Calvanese, V.; Verdin, E. Reactivation of latent hiv by histone deacetylase inhibitors. Trends Microbiol. 2013, 21, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valera, M.-S.; de Armas-Rillo, L.; Barroso-González, J.; Ziglio, S.; Batisse, J.; Dubois, N.; Marrero-Hernández, S.; Borel, S.; García-Expósito, L.; Biard-Piechaczyk, M. The hdac6/apobec3g complex regulates hiv-1 infectiveness by inducing vif autophagic degradation. Retrovirology 2015, 12, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Peng, L.; Seto, E.; Huang, S.; Qiu, Y. Modulation of histone deacetylase 6 (hdac6) nuclear import and tubulin deacetylase activity through acetylation. J. Biol. Chem. 2012, 287, 29168–29174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, X.; Ao, Z.; Trajtman, A.; Xu, W.; Kobinger, G.; Keynan, Y.; Yao, X. Hiv-1 envelope glycoprotein stimulates viral transcription and increases the infectivity of the progeny virus through the manipulation of cellular machinery. Sci. Rep. 2017, 7, 9487. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.J.; Liu, J.; Bertos, N.R.; Yang, X.-J. Identification of hdac10, a novel class ii human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002, 30, 1114–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, H.-Y.; Lee, C.-H.; Komarov, A.; Han, C.C.; Evans, R.M. Isolation and characterization of mammalian hdac10, a novel histone deacetylase. J. Biol. Chem. 2002, 277, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Kotian, S.; Liyanarachchi, S.; Zelent, A.; Parvin, J.D. Histone deacetylases 9 and 10 are required for homologous recombination. J. Biol. Chem. 2011, 286, 7722–7726. [Google Scholar] [CrossRef] [Green Version]
- Oehme, I.; Linke, J.-P.; Böck, B.C.; Milde, T.; Lodrini, M.; Hartenstein, B.; Wiegand, I.; Eckert, C.; Roth, W.; Kool, M. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc. Natl. Acad. Sci. USA 2013, 110, E2592–E2601. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Peng, L.; Seto, E. Histone deacetylase 10 regulates the cell cycle g2/m phase transition via a novel let-7–hmga2–cyclin a2 pathway. Mol. Cell. Biol. 2015, 35, 3547–3565. [Google Scholar] [CrossRef] [Green Version]
- Guardiola, A.R.; Yao, T.-P. Molecular cloning and characterization of a novel histone deacetylase hdac10. J. Biol. Chem. 2002, 277, 3350–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Huang, Y.; Wang, Z.; Wang, H.-T.; Duan, B.; Ye, D.; Wang, C.; Jing, R.; Leng, Y.; Xi, J. Hdac10 promotes lung cancer proliferation via akt phosphorylation. Oncotarget 2016, 7, 59388–59401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, F.P.; Tang, Q. Identification of cellular proteins that interact with human cytomegalovirus immediate-early protein 1 by protein array assay. Viruses 2013, 6, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R. The molecular biology of hiv integrase. Future Virol. 2012, 7, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Al-Mawsawi, L.Q.; Sechi, M.; Neamati, N. Single amino acid substitution in hiv-1 integrase catalytic core causes a dramatic shift in inhibitor selectivity. FEBS Lett. 2007, 581, 1151–1156. [Google Scholar] [CrossRef] [Green Version]
- Chiu, T.K.; Davies, D.R. Structure and function of hiv-1 integrase. Curr. Top. Med. Chem. 2004, 4, 965–977. [Google Scholar] [CrossRef]
- Engelman, A. Host cell factors and hiv-1 integration. Future HIV Ther. 2007, 1, 415–426. [Google Scholar] [CrossRef]
- Craigie, R.; Bushman, F.D. Host factors in retroviral integration and the selection of integration target sites. Microbiol. Spectr. 2014, 2, 1035–1050. [Google Scholar]
- Taltynov, O.; Desimmie, B.A.; Demeulemeester, J.; Christ, F.; Debyser, Z. Cellular cofactors of lentiviral integrase: From target validation to drug discovery. Mol. Biol. Int. 2012, 2012, 16. [Google Scholar] [CrossRef]
- Parissi, V.; Calmels, C.; De Soultrait, V.R.; Caumont, A.; Fournier, M.; Chaignepain, S.; Litvak, S. Functional interactions of human immunodeficiency virus type 1 integrase with human and yeast hsp60. J. Virol. 2001, 75, 11344–11353. [Google Scholar] [CrossRef] [Green Version]
- Kessl, J.J.; Kutluay, S.B.; Townsend, D.; Rebensburg, S.; Slaughter, A.; Larue, R.C.; Shkriabai, N.; Bakouche, N.; Fuchs, J.R.; Bieniasz, P.D. Hiv-1 integrase binds the viral rna genome and is essential during virion morphogenesis. Cell 2016, 166, 1257–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukovsky, A.; Göttlinger, H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 1996, 70, 6820–6825. [Google Scholar] [PubMed]
- Jurado, K.A.; Wang, H.; Slaughter, A.; Feng, L.; Kessl, J.J.; Koh, Y.; Wang, W.; Ballandras-Colas, A.; Patel, P.A.; Fuchs, J.R. Allosteric integrase inhibitor potency is determined through the inhibition of hiv-1 particle maturation. Proc. Natl. Acad. Sci. USA 2013, 110, 8690–8695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, M.J.; Monel, B.; Krishnan, L.; Shun, M.-C.; Di Nunzio, F.; Helland, D.E.; Engelman, A. Biochemical and virological analysis of the 18-residue c-terminal tail of hiv-1 integrase. Retrovirology 2009, 6, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelman, A. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 1999, 52, 411–426. [Google Scholar]
- Christ, F.; Shaw, S.; Demeulemeester, J.; Desimmie, B.A.; Marchand, A.; Butler, S.; Smets, W.; Chaltin, P.; Westby, M.; Debyser, Z. Small molecule inhibitors of the ledgf/p75 binding site of integrase (ledgins) block hiv replication and modulate integrase multimerization. Antimicrob. Agents Chemother. 2012, 56, 4365–4374. [Google Scholar] [CrossRef] [Green Version]
- Jayappa, K.D.; Ao, Z.; Wang, X.; Mouland, A.J.; Shekhar, S.; Yang, X.; Yao, X. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J. Virol. 2015, 89, 3497–3511. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.-J.; Mouland, A.J.; Subbramanian, R.A.; Forget, J.; Rougeau, N.; Bergeron, D.; Cohen, E.A. Vpr stimulates viral expression and induces cell killing in human immunodeficiency virus type 1-infected dividing jurkat t cells. J. Virol. 1998, 72, 4686–4693. [Google Scholar]
- Li, S.; Liu, C.; Klimov, A.; Subbarao, K.; Perdue, M.L.; Mo, D.; Ji, Y.; Woods, L.; Hietala, S.; Bryant, M. Recombinant influenza a virus vaccines for the pathogenic human a/hong kong/97 (h5n1) viruses. J. Infect. Dis. 1999, 179, 1132–1138. [Google Scholar] [CrossRef] [Green Version]
- Wahl-Jensen, V.; Kurz, S.K.; Hazelton, P.R.; Schnittler, H.-J.; Ströher, U.; Burton, D.R.; Feldmann, H. Role of ebola virus secreted glycoproteins and virus-like particles in activation of human macrophages. J. Virol. 2005, 79, 2413–2419. [Google Scholar] [CrossRef] [Green Version]
- Fischle, W.; Emiliani, S.; Hendzel, M.J.; Nagase, T.; Nomura, N.; Voelter, W.; Verdin, E. A new family of human histone deacetylases related tosaccharomyces cerevisiae hda1p. J. Biol. Chem. 1999, 274, 11713–11720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ao, Z.; Jayappa, K.D.; Wang, B.; Zheng, Y.; Kung, S.; Rassart, E.; Depping, R.; Kohler, M.; Cohen, E.A.; Yao, X. Importin α3 interacts with hiv-1 integrase and contributes to hiv-1 nuclear import and replication. J. Virol. 2010, 84, 8650–8663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ao, Z.; Huang, G.; Yao, H.; Xu, Z.; Labine, M.; Cochrane, A.W.; Yao, X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 2007, 282, 13456–13467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ao, Z.; Zhu, R.; Tan, X.; Liu, L.; Chen, L.; Liu, S.; Yao, X. Activation of hiv-1 expression in latently infected cd4+ t cells by the small molecule pkc412. Virol. J. 2016, 13, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ao, Z.; Jayappa, K.D.; Wang, B.; Zheng, Y.; Wang, X.; Peng, J.; Yao, X. Contribution of host nucleoporin 62 in hiv-1 integrase chromatin association and viral DNA integration. J. Biol. Chem. 2012, 287, 10544–10555. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Ao, Z.; Wang, B.; Jayappa, K.D.; Yao, X. Host protein ku70 binds and protects hiv-1 integrase from proteasomal degradation and is required for hiv replication. J. Biol. Chem. 2011, 286, 17722–17735. [Google Scholar] [CrossRef] [Green Version]
- Craigie, R. Hiv integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 2001, 276, 23213–23216. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Ao, Z.; Jayappa, K.D.; Yao, X. Characterization of the hiv-1 integrase chromatin-and ledgf/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase. Virol. J. 2010, 7, 68. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wei, S.-Q.; Engelman, A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type i intasome. J. Biol. Chem. 1999, 274, 17358–17364. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Yao, X. Posttranslational modifications of hiv-1 integrase by various cellular proteins during viral replication. Viruses 2013, 5, 1787–1801. [Google Scholar] [CrossRef] [Green Version]
- Cereseto, A.; Manganaro, L.; Gutierrez, M.I.; Terreni, M.; Fittipaldi, A.; Lusic, M.; Marcello, A.; Giacca, M. Acetylation of hiv-1 integrase by p300 regulates viral integration. EMBO J. 2005, 24, 3070–3081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terreni, M.; Valentini, P.; Liverani, V.; Gutierrez, M.I.; Di Primio, C.; Di Fenza, A.; Tozzini, V.; Allouch, A.; Albanese, A.; Giacca, M. Gcn5-dependent acetylation of hiv-1 integrase enhances viral integration. Retrovirology 2010, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuteja, R.; Tuteja, N. Ku autoantigen: A multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Downs, J.A.; Jackson, S.P. A means to a DNA end: The many roles of ku. Nat. Rev. Mol. Cell Biol. 2004, 5, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Giffin, W.; Torrance, H.; Rodda, D.J.; Préfontaine, G.G.; Pope, L.; Haché, R.J. Sequence-specific DNA binding by ku autoantigen and its effects on transcription. Nature 1996, 380, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clercq, E.; Debyser, Z. Hiv-1 integrase forms stable tetramers and associates with ledgf/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Hare, S.; Cherepanov, P. The interaction between lentiviral integrase and ledgf: Structural and functional insights. Viruses 2009, 1, 780–801. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, Y.; McClure, M.O.; Pizzato, M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol. 2008, 82, 12585–12588. [Google Scholar] [CrossRef] [Green Version]
- Malim, M.H.; Bieniasz, P.D. Hiv restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2012, 2, a006940. [Google Scholar] [CrossRef]
- He, Z.; Zhang, W.; Chen, G.; Xu, R.; Yu, X.-F. Characterization of conserved motifs in hiv-1 vif required for apobec3g and apobec3f interaction. J. Mol. Biol. 2008, 381, 1000–1011. [Google Scholar] [CrossRef]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The interferon-induced protein bst-2 restricts hiv-1 release and is downregulated from the cell surface by the viral vpu protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchand, C.; Johnson, A.A.; Semenova, E.; Pommier, Y. Mechanisms and inhibition of hiv integration. Drug Discov. Today Dis. Mech. 2006, 3, 253–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Maele, B.; Busschots, K.; Vandekerckhove, L.; Christ, F.; Debyser, Z. Cellular co-factors of hiv-1 integration. Trends Biochem. Sci. 2006, 31, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Allouch, A.; Di Primio, C.; Alpi, E.; Lusic, M.; Arosio, D.; Giacca, M.; Cereseto, A. The trim family protein kap1 inhibits hiv-1 integration. Cell Host Microbe 2011, 9, 484–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, Y.; Shinsky, S.A.; Porter, N.J.; Christianson, D.W. Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat. Commun. 2017, 8, 15368. [Google Scholar] [CrossRef] [Green Version]
- Poeschla, E.M. Integrase, ledgf/p75 and hiv replication. Cell. Mol. Life Sci. 2008, 65, 1403–1424. [Google Scholar] [CrossRef] [Green Version]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An essential role for ledgf/p75 in hiv integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for ledgf/p75 in targeting hiv DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef]
- Lever, A.M.; Jeang, K.-T. Insights into cellular factors that regulate hiv-1 replication in human cells. Biochemistry 2011, 50, 920–931. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, C. Bst-2 diminishes hiv-1 infectivity. J. Virol. 2010, 84, 12336–12343. [Google Scholar] [CrossRef] [Green Version]
- Le Rouzic, E.; Bonnard, D.; Chasset, S.; Bruneau, J.-M.; Chevreuil, F.; Le Strat, F.; Nguyen, J.; Beauvoir, R.; Amadori, C.; Brias, J. Dual inhibition of hiv-1 replication by integrase-ledgf allosteric inhibitors is predominant at the post-integration stage. Retrovirology 2013, 10, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desimmie, B.A.; Schrijvers, R.; Demeulemeester, J.; Borrenberghs, D.; Weydert, C.; Thys, W.; Vets, S.; Van Remoortel, B.; Hofkens, J.; De Rijck, J. Ledgins inhibit late stage hiv-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, X.; Ao, Z.; Olukitibi, T.; Yao, X. Characterization of the Role of Host Cellular Factor Histone Deacetylase 10 during HIV-1 Replication. Viruses 2020, 12, 28. https://0-doi-org.brum.beds.ac.uk/10.3390/v12010028
Ran X, Ao Z, Olukitibi T, Yao X. Characterization of the Role of Host Cellular Factor Histone Deacetylase 10 during HIV-1 Replication. Viruses. 2020; 12(1):28. https://0-doi-org.brum.beds.ac.uk/10.3390/v12010028
Chicago/Turabian StyleRan, Xiaozhuo, Zhujun Ao, Titus Olukitibi, and Xiaojian Yao. 2020. "Characterization of the Role of Host Cellular Factor Histone Deacetylase 10 during HIV-1 Replication" Viruses 12, no. 1: 28. https://0-doi-org.brum.beds.ac.uk/10.3390/v12010028