Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies
Abstract
:1. Introduction
2. Solid-State NMR
3. NMR Sample Preparation
3.1. The Need for Recombinant Sample Production
3.2. The Size of the Proteins and Their Assemblies Amenable to “High Resolution” Solid-State NMR
3.3. Labeled Protein Production
4. NMR Protocols
4.1. Sequential Resonance Assignments
4.2. Chemical Shifts Reveal Structural Information
4.3. Structure Determination
4.4. Investigating Interactions
5. Examples of Solid-State NMR Studies on Viral Proteins
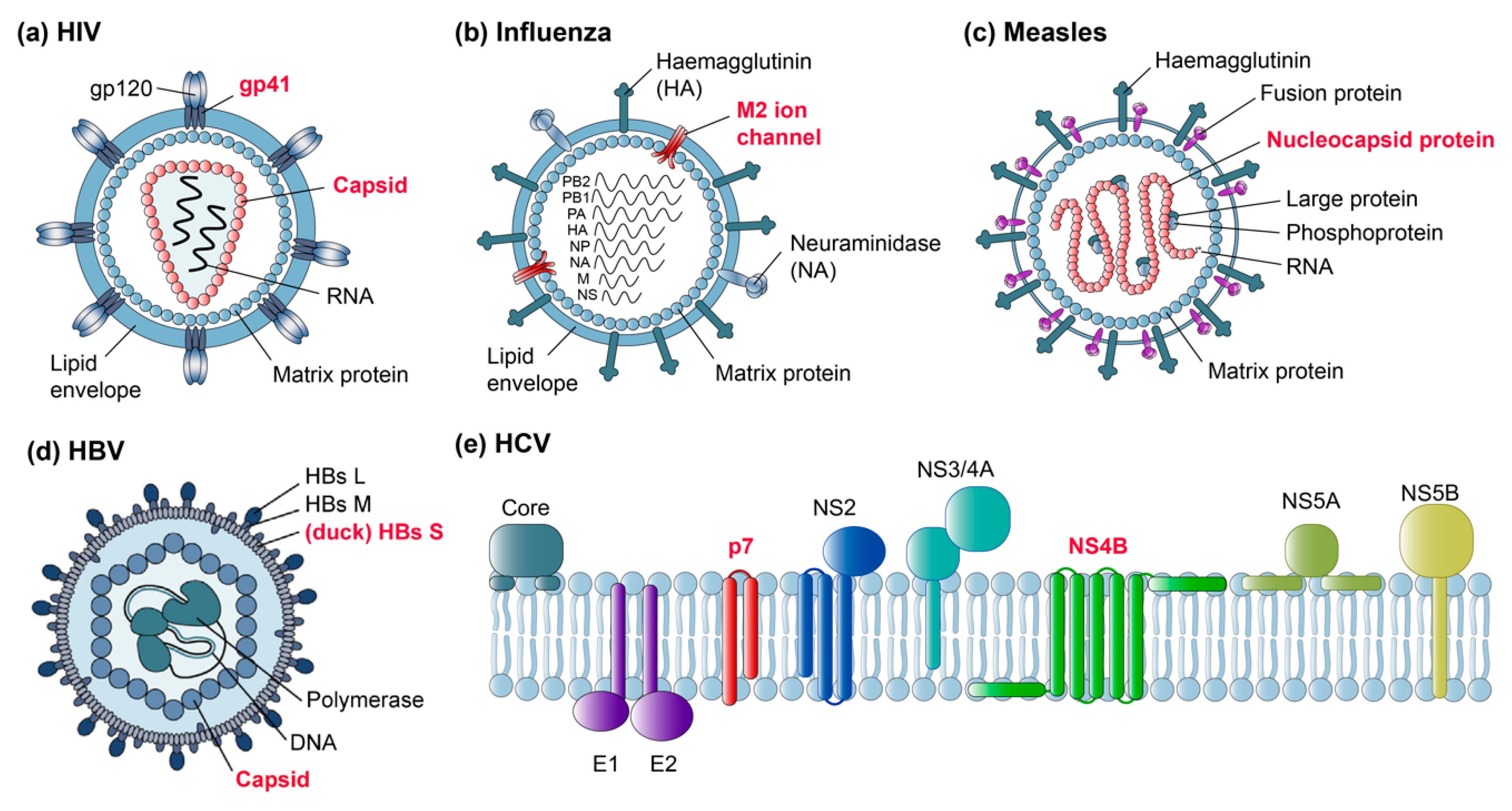
5.1. Viral Capsids
5.1.1. HIV Capsids
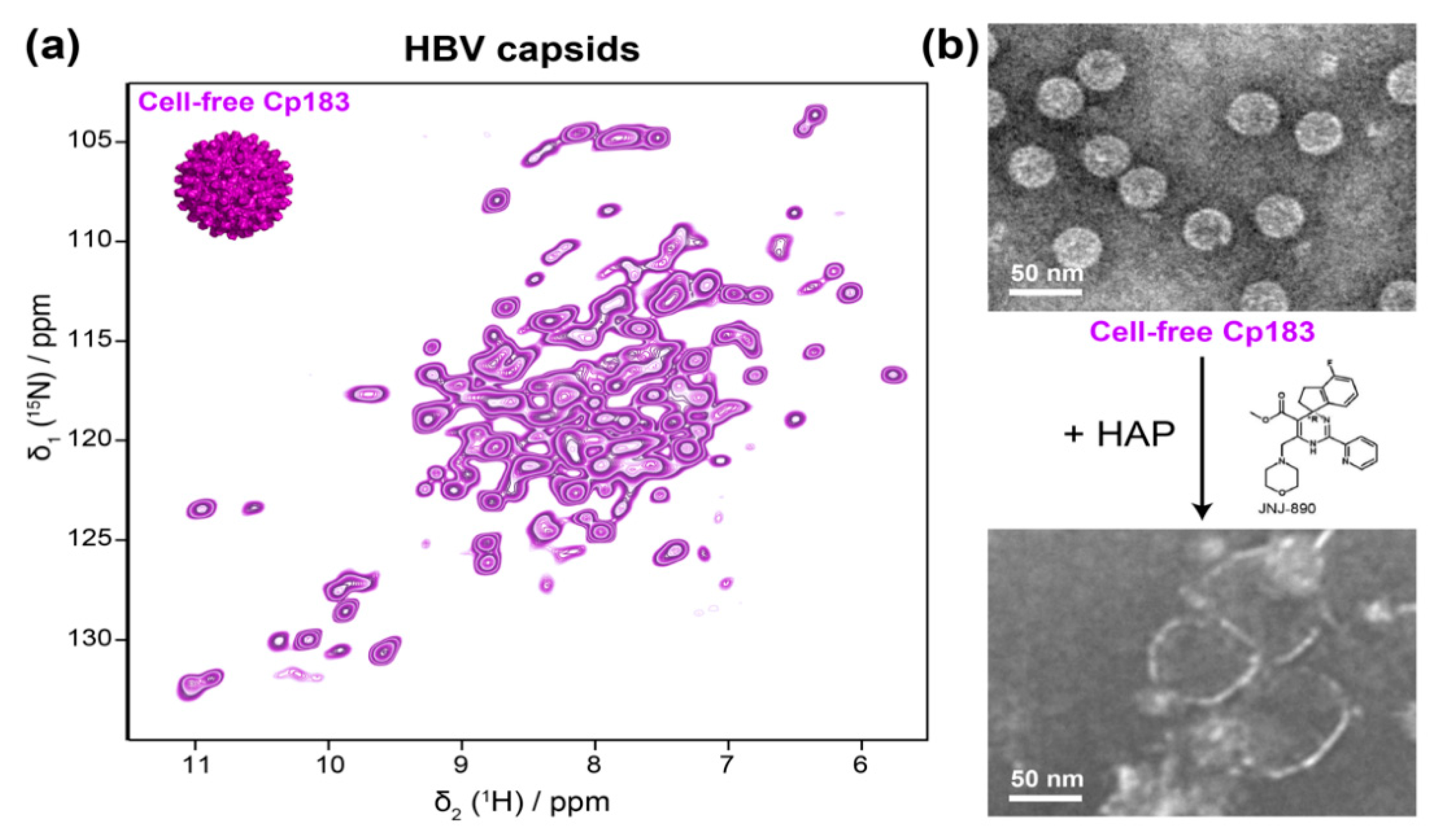
5.1.2. HBV Capsids
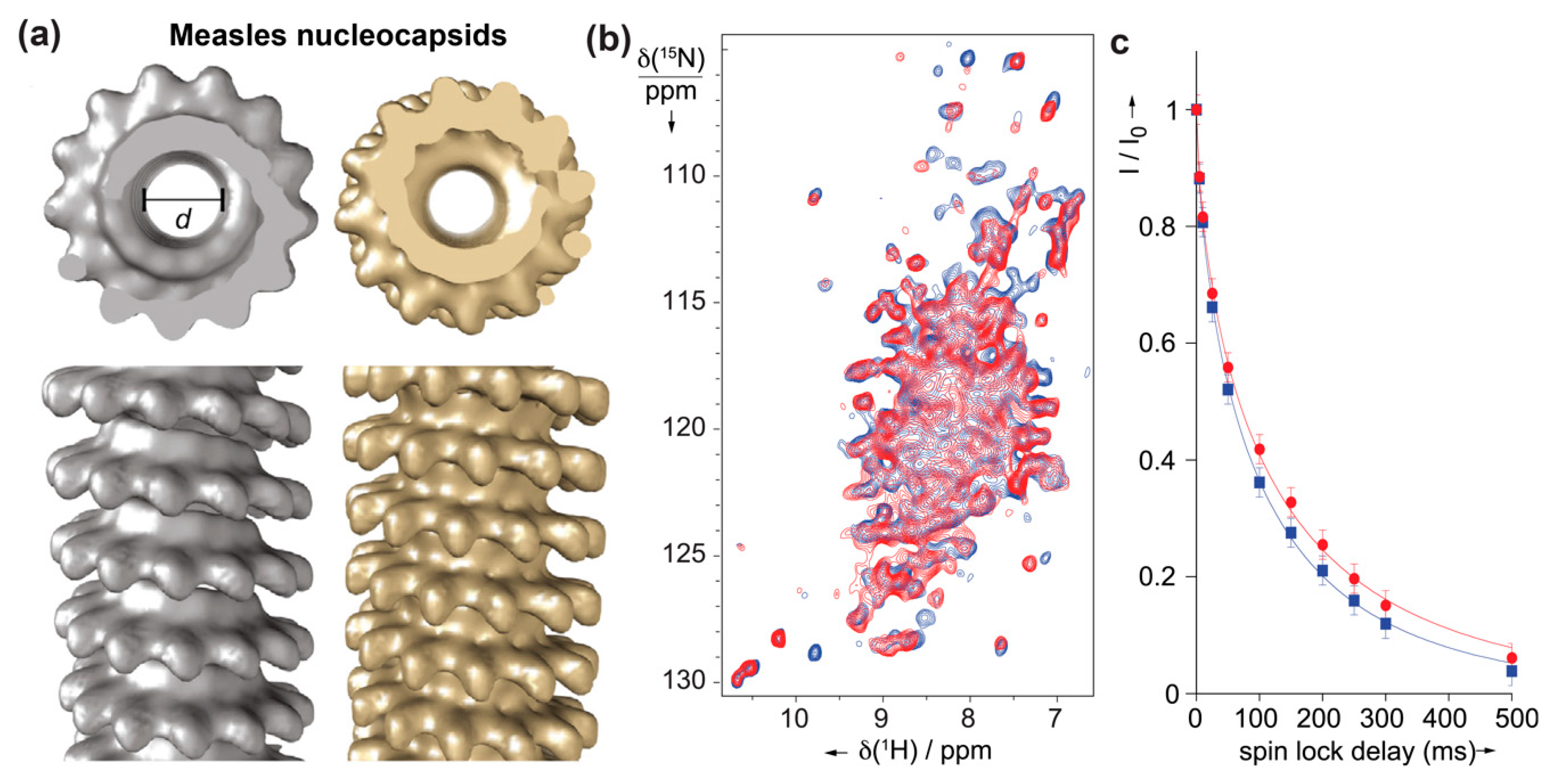
5.1.3. Measles Virus Nucleocapsids
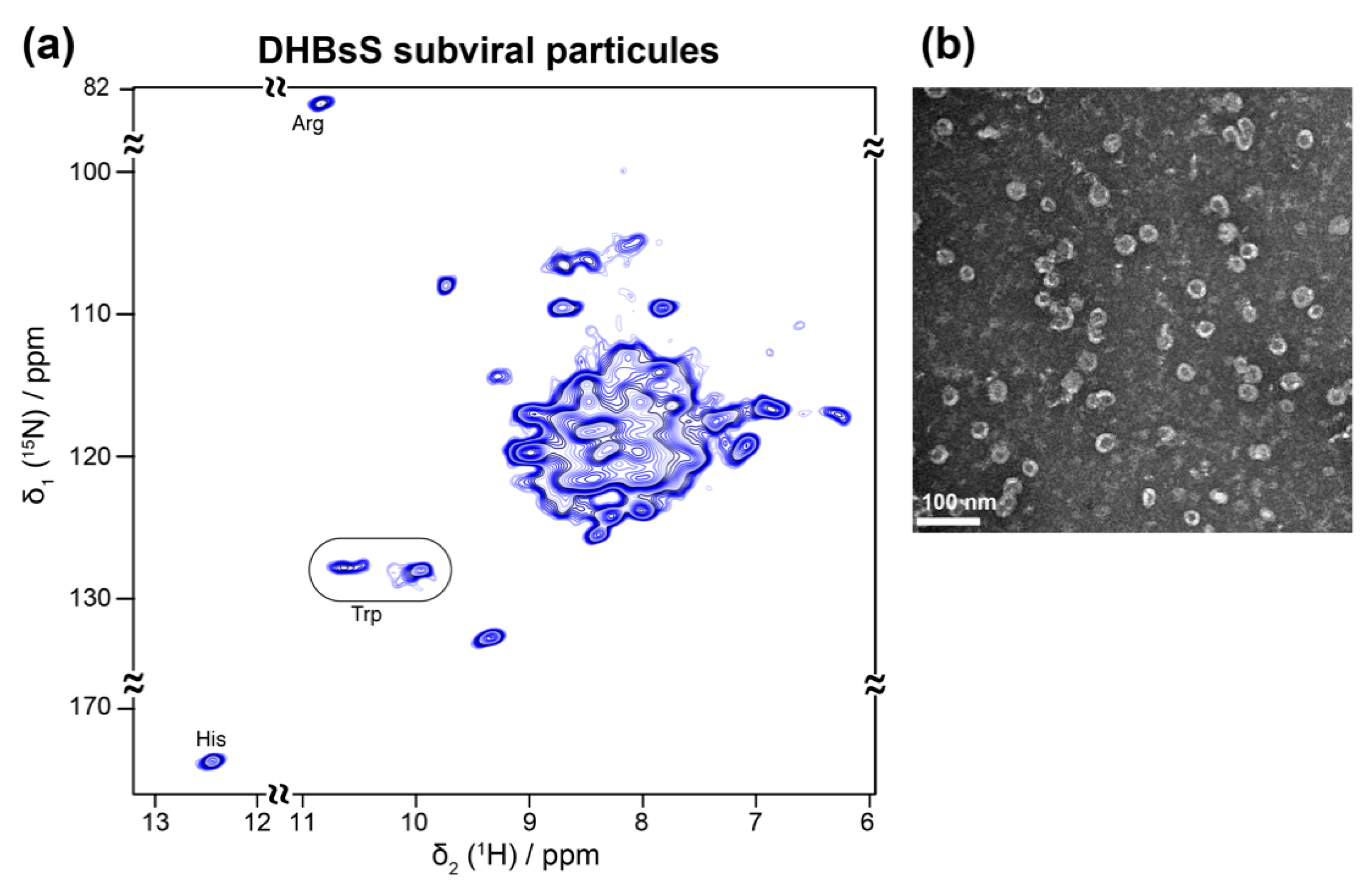
5.2. Viral Envelopes
5.3. Other Viral Membrane Proteins
5.3.1. HIV gp41 and Vpu
5.3.2. Influenza M2 Channel
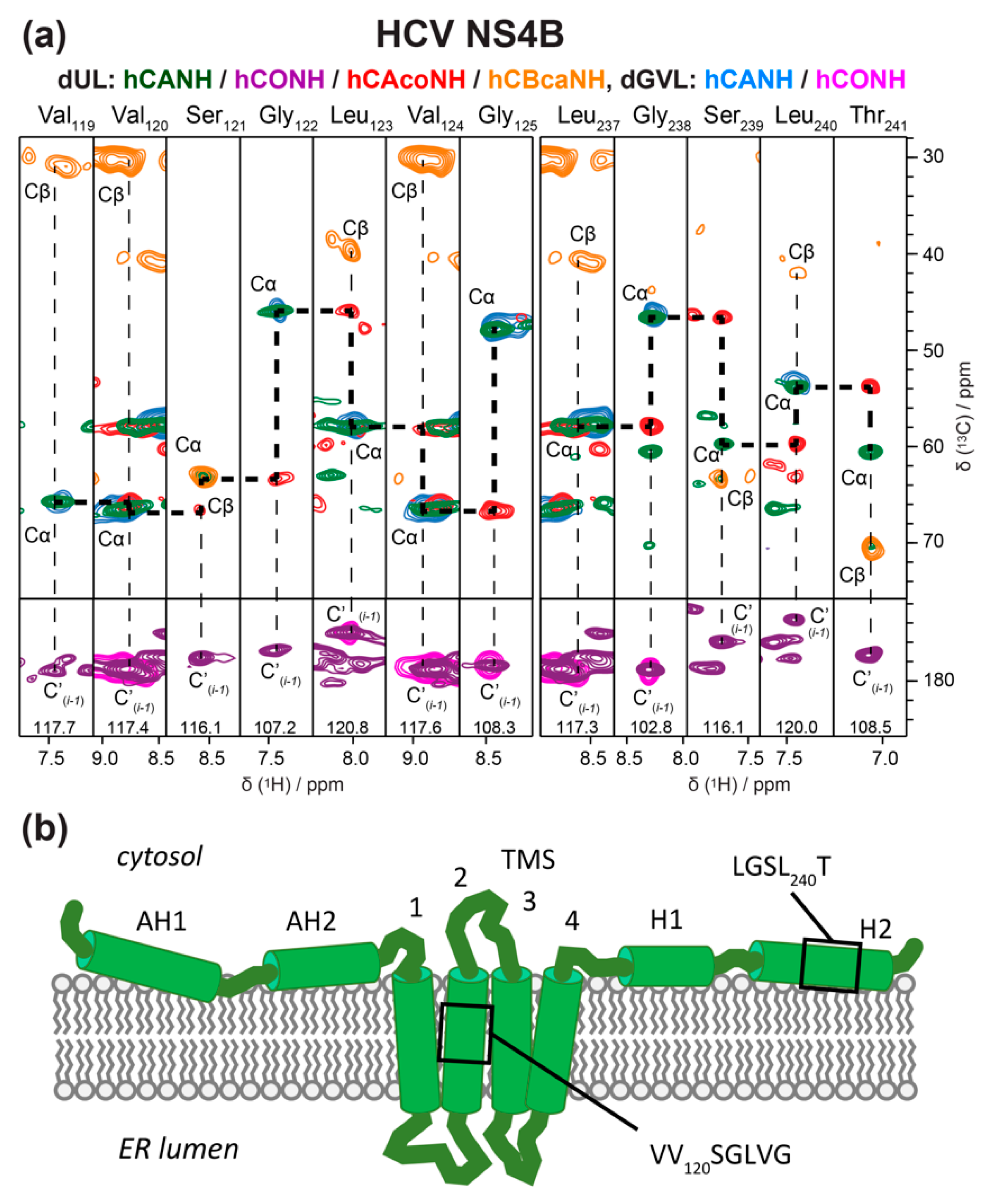
5.3.3. HCV p7 and NS4B
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Hopper, P.; Harrison, S.C.; Sauer, R.T. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J. Mol. Biol. 1984, 177, 701–713. [Google Scholar] [CrossRef]
- Silva, A.M.; Rossmann, M.G. Refined structure of southern bean mosaic virus at 2.9 A resolution. J. Mol. Biol. 1987, 197, 69–87. [Google Scholar] [CrossRef]
- Krishnaswamy, S.; Rossmann, M.G. Structural refinement and analysis of Mengo virus. J. Mol. Biol. 1990, 211, 803–844. [Google Scholar] [CrossRef]
- Arnold, E.; Rossmann, M.G. The use of molecular-replacement phases for the refinement of the human rhinovirus 14 structure. Acta Crystallogr. A Found. Crystallogr. 1988, 44, 270–282. [Google Scholar] [CrossRef]
- Kim, S.S.; Smith, T.J.; Chapman, M.S.; Rossmann, M.C.; Pevear, D.C.; Dutko, F.J.; Felock, P.J.; Diana, G.D.; McKinlay, M.A. Crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 1989, 210, 91–111. [Google Scholar] [CrossRef]
- Filman, D.J.; Syed, R.; Chow, M.; Macadam, A.J.; Minor, P.D.; Hogle, J.M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989, 8, 1567–1579. [Google Scholar] [CrossRef] [PubMed]
- Wynne, S.A.; Crowther, R.A.; Leslie, A.G. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 1999, 3, 771–780. [Google Scholar] [CrossRef]
- Elias, M.; Liebschner, D.; Koepke, J.; Lecomte, C.; Guillot, B.; Jelsch, C.; Chabriere, E. Hydrogen atoms in protein structures: High-resolution X-ray diffraction structure of the DFPase. BMC Res. Notes 2013, 6, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woińska, M.; Grabowsky, S.; Dominiak, P.M.; Woźniak, K.; Jayatilaka, D. Hydrogen atoms can be located accurately and precisely by x-ray crystallography. Sci. Adv. 2016, 2, e1600192. [Google Scholar] [CrossRef] [Green Version]
- Packianathan, C.; Katen, S.P.; Dann, C.E.; Zlotnick, A. Conformational Changes in the Hepatitis B Virus Core Protein Are Consistent with a Role for Allostery in Virus Assembly. J. Virol. 2010, 84, 1607–1615. [Google Scholar] [CrossRef] [Green Version]
- Klumpp, K.; Lam, A.M.; Lukacs, C.; Vogel, R.; Ren, S.; Espiritu, C.; Baydo, R.; Atkins, K.; Abendroth, J.; Liao, G.; et al. High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc. Natl. Acad. Sci. USA 2015, 112, 15196–15201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Guu, T.S.Y.; Cao, J.; Li, Y.; Cheng, L.; Tao, Y.J.; Zhang, J. Structure determination of a human virus by the combination of cryo-EM and X-ray crystallography. Biophys Rep. 2016, 2, 55–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttcher, B.; Wynne, S.A.; Crowther, R.A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 1997, 386, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.F.; Cheng, N.; Zlotnick, A.; Wingfield, P.T.; Stahl, S.J.; Steven, A.C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 1997, 386, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, B.; Nassal, M. Structure of Mutant Hepatitis B Core Protein Capsids with Premature Secretion Phenotype. J. Mol. Biol. 2018, 430, 4941–4954. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhu, D.; Agarkova, I.; Adhikari, J.; Klose, T.; Liu, Y.; Chen, Z.; Sun, Y.; Gross, M.L.; Van Etten, J.L.; et al. Near-atomic structure of a giant virus. Nat. Commun. 2019, 10, 388. [Google Scholar] [CrossRef] [Green Version]
- Makbul, C.; Nassal, M.; Böttcher, B. Slowly folding surface extension in the prototypic avian hepatitis B virus capsid governs stability. eLife 2020, 9. [Google Scholar] [CrossRef]
- Bloch, F.; Hansen, W.W.; Packard, M. The Nuclear Induction Experiment. Phys. Rev. 1946, 70, 474–485. [Google Scholar] [CrossRef]
- Purcell, E.M.; Torrey, H.C.; Pound, R.V. Resonance Absorption by Nuclear Magnetic Moments in a Solid. Phys. Rev. 1946, 69, 37–38. [Google Scholar] [CrossRef]
- Aue, W.P.; Bartholdi, E.; Ernst, R.R. Two-dimensional spectroscopy. Application to nuclear magnetic resonance. J. Chem. Phys. 1976, 64, 2229–2246. [Google Scholar] [CrossRef] [Green Version]
- Jeener, J.; Meier, B.H.; Bachmann, P.; Ernst, R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979, 71, 4546–4553. [Google Scholar] [CrossRef]
- Jeener, J.; Alewaeters, G. “Pulse pair technique in high resolution NMR” a reprint of the historical 1971 lecture notes on two-dimensional spectroscopy. Prog. Nucl. Mag. Res. Spectrosc. 2016, 94–95, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR studies of structure and function of biological macromolecules (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2003, 42, 3340–3363. [Google Scholar] [CrossRef] [PubMed]
- Religa, T.L.; Ruschak, A.M.; Rosenzweig, R.; Kay, L.E. Site-directed methyl group labeling as an NMR probe of structure and dynamics in supramolecular protein systems: Applications to the proteasome and to the ClpP protease. J. Am. Chem. Soc. 2011, 133, 9063–9068. [Google Scholar] [CrossRef]
- Andrew, E.R.; Bradbury, A.; Eades, R.G. Removal of Dipolar Broadening of Nuclear Magnetic Resonance Spectra of Solids by Specimen Rotation. Nature 1959, 183, 1802–1803. [Google Scholar] [CrossRef]
- Zhang, Z.; Oss, A.; Org, M.-L.; Samoson, A.; Li, M.; Tan, H.; Su, Y.; Yang, J. Selectively Enhanced 1H-1H Correlations in Proton-Detected Solid-State NMR under Ultrafast MAS Conditions. J. Phys. Chem. Lett. 2020, 11, 8077–8083. [Google Scholar] [CrossRef]
- Schledorn, M.; Malär, A.A.; Torosyan, A.; Penzel, S.; Klose, D.; Oss, A.; Org, M.-L.; Wang, S.; Lecoq, L.; Cadalbert, R.; et al. Protein NMR Spectroscopy at 150 kHz Magic-Angle Spinning Continues To Improve Resolution and Mass Sensitivity. ChemBioChem. Biol. 2020, 115, 11519. [Google Scholar] [CrossRef]
- McDermott, A.E.; Polenova, T.; Böckmann, A.; Zilm, K.W.; Paulsen, E.K.; Martin, R.W.; Montelione, G.T. Partial NMR assignments for uniformly (13C, 15N)-enriched BPTI in the solid state. J. Biomol. NMR 2000, 16, 209–219. [Google Scholar] [CrossRef]
- Castellani, F.; van Rossum, B.; Diehl, A.; Schubert, M.; Rehbein, K.; Oschkinat, H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 2002, 420, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Linser, R.; Bardiaux, B.; Andreas, L.B.; Hyberts, S.G.; Morris, V.K.; Pintacuda, G.; Sunde, M.; Kwan, A.H.; Wagner, G. Solid-State NMR Structure Determination from Diagonal-Compensated, Sparsely Nonuniform-Sampled 4D Proton–Proton Restraints. J. Am. Chem. Soc. 2014, 136, 11002–11010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbet-Massin, E.; Pell, A.J.; Retel, J.S.; Andreas, L.B.; Jaudzems, K.; Franks, W.T.; Nieuwkoop, A.J.; Hiller, M.; Higman, V.; Guerry, P.; et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 2014, 136, 12489–12497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, V.; Penzel, S.; Szekely, K.; Cadalbert, R.; Testori, E.; Oss, A.; Past, J.; Samoson, A.; Ernst, M.; Böckmann, A.; et al. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew. Chem. Int. Ed. 2014, 53, 12253–12256. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, L.; Schledorn, M.; Wang, S.; Smith-Penzel, S.; Malär, A.A.; Callon, M.; Nassal, M.; Meier, B.H.; Böckmann, A. 100 kHz MAS Proton-Detected NMR Spectroscopy of Hepatitis B Virus Capsids. Front. Mol. Biosci. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Penzel, S.; Oss, A.; Org, M.-L.; Samoson, A.; Böckmann, A.; Ernst, M.; Meier, B.H. Spinning faster: Protein NMR at MAS frequencies up to 126 kHz. J. Biomol. NMR 2019, 73, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiennet, C.; Schütz, A.K.; Hunkeler, A.; Kunert, B.; Terradot, L.; Böckmann, A.; Meier, B.H. A Sedimented Sample of a 59 kDa Dodecameric Helicase Yields High-Resolution Solid-State NMR Spectra. Angew. Chem. Int. Ed. 2012, 51, 7855–7858. [Google Scholar] [CrossRef]
- Bertini, I.; Luchinat, C.; Parigi, G.; Ravera, E.; Reif, B.; Turano, P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc. Natl. Acad. Sci. USA 2011, 108, 10396–10399. [Google Scholar] [CrossRef] [Green Version]
- Andreas, L.B.; Jaudzems, K.; Stanek, J.; Lalli, D.; Bertarello, A.; Le Marchand, T.; Cala-De Paepe, D.; Kotelovica, S.; Akopjana, I.; Knott, B.; et al. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc. Natl. Acad. Sci. USA 2016, 113, 9187–9192. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Fogeron, M.-L.; Schledorn, M.; Dujardin, M.; Penzel, S.; Burdette, D.; Berke, J.M.; Nassal, M.; Lecoq, L.; Meier, B.H.; et al. Combining Cell-Free Protein Synthesis and NMR Into a Tool to Study Capsid Assembly Modulation. Front. Mol. Biosci. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- David, G.; Fogeron, M.-L.; Schledorn, M.; Montserret, R.; Haselmann, U.; Penzel, S.; Badillo, A.; Lecoq, L.; André, P.; Nassal, M.; et al. Structural Studies of Self-Assembled Subviral Particles: Combining Cell-Free Expression with 110 kHz MAS NMR Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 4787–4791. [Google Scholar] [CrossRef] [PubMed]
- Schanda, P.; Ernst, M. Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: Principles and applications to biomolecules. Prog. Nucl. Mag. Res. Sp. 2016, 96, 1–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasmer, C.; Lange, A.; Van Melckebeke, H.; Siemer, A.B.; Riek, R.; Meier, B.H. Amyloid Fibrils of the HET-s(218-289) Prion Form a Solenoid with a Triangular Hydrophobic Core. Science 2008, 319, 1523–1526. [Google Scholar] [CrossRef] [PubMed]
- Lacabanne, D.; Fogeron, M.-L.; Wiegand, T.; Cadalbert, R.; Meier, B.H.; Böckmann, A. Protein sample preparation for solid-state NMR investigations. Prog. Nucl. Mag. Res. Spectrosc. 2019, 110, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Thiriot, D.S.; Nevzorov, A.A.; Zagyanskiy, L.; Wu, C.H.; Opella, S.J. Structure of the Coat Protein in Pf1 Bacteriophage Determined by Solid-state NMR Spectroscopy. J. Mol. Biol. 2004, 341, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Goldbourt, A.; Gross, B.J.; Day, L.A.; McDermott, A.E. Filamentous Phage Studied by Magic-Angle Spinning NMR: Resonance Assignment and Secondary Structure of the Coat Protein in Pf1. J. Am. Chem. Soc. 2007, 129, 2338–2344. [Google Scholar] [CrossRef]
- Morag, O.; Abramov, G.; Goldbourt, A. Complete Chemical Shift Assignment of the ssDNA in the Filamentous Bacteriophage fd Reports on Its Conformation and on Its Interface with the Capsid Shell. J. Am. Chem. Soc. 2014, 136, 2292–2301. [Google Scholar] [CrossRef]
- Abramov, G.; Morag, O.; Goldbourt, A. Magic-angle spinning NMR of intact bacteriophages: Insights into the capsid, DNA and their interface. J. Magn. Reson. 2015, 253, 80–90. [Google Scholar] [CrossRef]
- Sergeyev, I.V.; Day, L.A.; Goldbourt, A.; McDermott, A.E. Chemical Shifts for the Unusual DNA Structure in Pf1 Bacteriophage from Dynamic-Nuclear-Polarization-Enhanced Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2011, 133, 20208–20217. [Google Scholar] [CrossRef]
- Sergeyev, I.V.; Itin, B.; Rogawski, R.; Day, L.A.; McDermott, A.E. Efficient assignment and NMR analysis of an intact virus using sequential side-chain correlations and DNP sensitization. Proc. Natl. Acad. Sci. USA 2017, 114, 5171–5176. [Google Scholar] [CrossRef] [Green Version]
- Maly, T.; Debelouchina, G.T.; Bajaj, V.S.; Hu, K.-N.; Joo, C.-G.; Mak-Jurkauskas, M.L.; Sirigiri, J.R.; van der Wel, P.C.A.; Herzfeld, J.; Temkin, R.J.; et al. Dynamic nuclear polarization at high magnetic fields. J. Chem. Phys. 2008, 128, 052211. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, T.; Hunkeler, A.; Däpp, A.; Verasdonck, J.; Cadalbert, R.; Bousset, L.; Melki, R.; Böckmann, A.; Meier, B.H. CONFINE-MAS: A magic-angle spinning NMR probe that confines the sample in case of a rotor explosion. J. Biomol. NMR 2018, 72, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Penzel, S.; Smith, A.A.; Agarwal, V.; Hunkeler, A.; Org, M.-L.; Samoson, A.; Böckmann, A.; Ernst, M.; Meier, B.H. Protein resonance assignment at MAS frequencies approaching 100 kHz: A quantitative comparison of J-coupling and dipolar-coupling-based transfer methods. J. Biomol. NMR 2015, 63, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Lacabanne, D.; Torosyan, A.; Boudet, J.; Cadalbert, R.; Allain, F.H.T.; Meier, B.H.; Böckmann, A. Sedimentation Yields Long-Term Stable Protein Samples as Shown by Solid-State NMR. Front. Mol. Biosci. 2020, 7, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecoq, L.; Wang, S.; Wiegand, T.; Bressanelli, S.; Nassal, M.; Meier, B.H.; Böckmann, A. Localizing Conformational Hinges by NMR: Where Do Hepatitis B Virus Core Proteins Adapt for Capsid Assembly? ChemPhysChem 2018, 19, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Saxena, K.; Schwalbe, H.; Klein-Seetharaman, J. Isotope labeling in mammalian cells. Methods Mol. Biol. 2012, 831, 55–69. [Google Scholar]
- Heger-Stevic, J.; Zimmermann, P.; Lecoq, L.; Böttcher, B.; Nassal, M. Hepatitis B virus core protein phosphorylation: Identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog 2018, 14, e1007488-31. [Google Scholar] [CrossRef] [Green Version]
- David, G.; Fogeron, M.-L.; Montserret, R.; Lecoq, L.; Page, A.; Delolme, F.; Nassal, M.; Böckmann, A. Phosphorylation and Alternative Translation on Wheat Germ Cell-Free Protein Synthesis of the DHBV Large Envelope Protein. Front. Mol. Biosci. 2019, 6, 138. [Google Scholar] [CrossRef]
- Fogeron, M.-L.; Jirasko, V.; Penzel, S.; Paul, D.; Montserret, R.; Danis, C.; Lacabanne, D.; Badillo, A.; Gouttenoire, J.; Moradpour, D.; et al. Cell-free expression, purification, and membrane reconstitution for NMR studies of the nonstructural protein 4B from hepatitis C virus. J. Biomol. NMR 2016, 65, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Jirasko, V.; Lakomek, N.-A.; Penzel, S.; Fogeron, M.-L.; Bartenschlager, R.; Meier, B.H.; Böckmann, A. Proton-Detected Solid-State NMR of the Cell-Free Synthesized α-Helical Transmembrane Protein NS4B from Hepatitis C Virus. ChemBioChem. Biol. 2020, 1, 5–9. [Google Scholar] [CrossRef]
- Morita, E.-H.; Sawasaki, T.; Tanaka, R.; Endo, Y.; Kohno, T. A wheat germ cell-free system is a novel way to screen protein folding and function. Protein Sci. 2003, 12, 1216–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, E.-H.; Shimizu, M.; Ogasawara, T.; Endo, Y.; Tanaka, R.; Kohno, T. A novel way of amino acid-specific assignment in (1)H-(15)N HSQC spectra with a wheat germ cell-free protein synthesis system. J. Biomol. NMR 2004, 30, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, M.; Singarapu, K.K.; Makino, S.-I.; Sahu, S.C.; Matsubara, Y.; Endo, Y.; Kainosho, M.; Markley, J.L. Hydrogen exchange during cell-free incorporation of deuterated amino acids and an approach to its inhibition. J. Biomol. NMR 2011, 51, 467–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevelkov, V.; Rehbein, K.; Diehl, A.; Reif, B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew. Chem. Int. Ed. Engl. 2006, 45, 3878–3881. [Google Scholar] [CrossRef] [PubMed]
- Asami, S.; Szekely, K.; Schanda, P.; Meier, B.H.; Reif, B. Optimal degree of protonation for 1H detection of aliphatic sites in randomly deuterated proteins as a function of the MAS frequency. J. Biomol. NMR 2012, 54, 155–168. [Google Scholar] [CrossRef]
- Kohno, T.; Endo, Y. Production of protein for nuclear magnetic resonance study using the wheat germ cell-free system. Methods Mol. Biol. 2007, 375, 257–272. [Google Scholar]
- Gupta, S.; Tycko, R. Segmental isotopic labeling of HIV-1 capsid protein assemblies for solid state NMR. J. Biomol. NMR 2018, 70, 103–114. [Google Scholar] [CrossRef]
- Gupta, S.; Louis, J.M.; Tycko, R. Effects of an HIV-1 maturation inhibitor on the structure and dynamics of CA-SP1 junction helices in virus-like particles. Proc. Natl. Acad. Sci. USA 2020, 117, 10286–10293. [Google Scholar] [CrossRef]
- Etzkorn, M.; Böckmann, A.; Lange, A.; Baldus, M. Probing molecular interfaces using 2D magic-angle-spinning NMR on protein mixtures with different uniform labeling. J. Am. Chem. Soc. 2004, 126, 14746–14751. [Google Scholar] [CrossRef]
- Lundström, P.; Teilum, K.; Carstensen, T.; Bezsonova, I.; Wiesner, S.; Hansen, D.F.; Religa, T.L.; Akke, M.; Kay, L.E. Fractional 13C enrichment of isolated carbons using [1-13C]- or [2- 13C]-glucose facilitates the accurate measurement of dynamics at backbone Calpha and side-chain methyl positions in proteins. J. Biomol. NMR 2007, 38, 199–212. [Google Scholar] [CrossRef]
- Loquet, A.; Habenstein, B.; Lange, A. Structural Investigations of Molecular Machines by Solid-State NMR. Acc. Chem. Res. 2013, 46, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Rienstra, C.M.; Hohwy, M.; Hong, M.; Griffin, R.G. 2D and 3D 15N− 13C− 13C NMR Chemical Shift Correlation Spectroscopy of Solids: Assignment of MAS Spectra of Peptides. J. Am. Chem. Soc. 2000, 122, 10979–10990. [Google Scholar] [CrossRef]
- Pauli, J.; Baldus, M.; van Rossum, B.; de Groot, H.; Oschkinat, H. Backbone and side-chain 13C and 15N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 Tesla. ChemBioChem. Biol. 2001, 2, 272–281. [Google Scholar] [CrossRef]
- Habenstein, B.; Wasmer, C.; Bousset, L.; Sourigues, Y.; Schütz, A.; Loquet, A.; Meier, B.H.; Melki, R.; Böckmann, A. Extensive de novo solid-state NMR assignments of the 33 kDa C-terminal domain of the Ure2 prion. J. Biomol. NMR 2011, 51, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuetz, A.; Wasmer, C.; Habenstein, B.; Verel, R.; Greenwald, J.; Riek, R.; Böckmann, A.; Meier, B.H. Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). ChemBioChem. Biol. 2010, 11, 1543–1551. [Google Scholar] [CrossRef]
- Porterfield, J.Z.; Dhason, M.S.; Loeb, D.D.; Nassal, M.; Stray, S.J.; Zlotnick, A. Full-Length Hepatitis B Virus Core Protein Packages Viral and Heterologous RNA with Similarly High Levels of Cooperativity. J. Virol. 2010, 84, 7174–7184. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Yao, Y.; Marassi, F.M. Membrane protein structure determination in membrana. Acc. Chem. Res. 2013, 46, 2182–2190. [Google Scholar] [CrossRef] [Green Version]
- Güntert, P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009, 38, 129–143. [Google Scholar] [CrossRef]
- Linge, J.P.; O’Donoghue, S.I.; Nilges, M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Meth. Enzymol. 2001, 339, 71–90. [Google Scholar]
- Bardiaux, B.; van Rossum, B.-J.; Nilges, M.; Oschkinat, H. Efficient modeling of symmetric protein aggregates from NMR data. Angew. Chem. Int. Ed. 2012, 51, 6916–6919. [Google Scholar] [CrossRef]
- Davis, B. Screening Protein–Small Molecule Interactions by NMR. In Protein NMR; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1008, pp. 389–413. [Google Scholar]
- Dias, D.M.; Ciulli, A. NMR approaches in structure-based lead discovery: Recent developments and new frontiers for targeting multi-protein complexes. Prog. Biophys. Mol. Biol. 2014, 116, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opella, S.J.; Zeri, A.C.; Park, S.H. Structure, Dynamics, and Assembly of Filamentous Bacteriophages by Nuclear Magnetic Resonance Spectroscopy. Annu. Rev. Phys. Chem. 2008, 59, 635–657. [Google Scholar] [CrossRef] [PubMed]
- Goldbourt, A. Structural characterization of bacteriophage viruses by NMR. Prog. Nucl. Mag. Res. Spectrosc. 2019, 114–115, 192–210. [Google Scholar] [CrossRef] [PubMed]
- Marassi, F.M. NMR of peptides and proteins in oriented membranes. Concepts Magn. Reson. 2002, 14, 212–224. [Google Scholar] [CrossRef]
- Urban, S.; Bartenschlager, R.; Kubitz, R.; Zoulim, F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology 2014, 147, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Bartenschlager, R.; Lohmann, V.; Penin, F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013, 11, 482–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Hill, C.P.; Sundquist, W.I.; Finch, J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 2000, 407, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.M.; Lu, M.; Suiter, C.L.; Hou, G.; Zhang, H.; Polenova, T. Magic angle spinning NMR of viruses. Prog. Nucl. Mag. Res. Sp. 2015, 86–87, 21–40. [Google Scholar] [CrossRef] [Green Version]
- Suiter, C.L.; Quinn, C.M.; Lu, M.; Hou, G.; Zhang, H.; Polenova, T. MAS NMR of HIV-1 protein assemblies. J. Magn. Reson. 2015, 253, 10–22. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Ahn, J.; Concel, J.; Byeon, I.-J.L.; Gronenborn, A.M.; Yang, J.; Polenova, T. Solid-State NMR Studies of HIV-1 Capsid Protein Assemblies. J. Am. Chem. Soc. 2010, 132, 1976–1987. [Google Scholar] [CrossRef] [Green Version]
- Byeon, I.-J.L.; Hou, G.; Han, Y.; Suiter, C.L.; Ahn, J.; Jung, J.; Byeon, C.-H.; Gronenborn, A.M.; Polenova, T. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: Implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 2012, 134, 6455–6466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Hou, G.; Zhang, H.; Suiter, C.L.; Ahn, J.; Byeon, I.-J.L.; Perilla, J.R.; Langmead, C.J.; Hung, I.; Gor’kov, P.L.; et al. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc. Natl. Acad. Sci. USA 2015, 112, 14617–14622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hou, G.; Lu, M.; Ahn, J.; Byeon, I.-J.L.; Langmead, C.J.; Perilla, J.R.; Hung, I.; Gor’kov, P.L.; Gan, Z.; et al. HIV-1 Capsid Function is Regulated by Dynamics: Quantitative Atomic-Resolution Insights by Integrating Magic-Angle-Spinning NMR, QM/MM, and MD. J. Am. Chem. Soc. 2016, 138, 14066–14075. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.M.; Wang, M.; Fritz, M.P.; Runge, B.; Ahn, J.; Xu, C.; Perilla, J.R.; Gronenborn, A.M.; Polenova, T. Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2018, 115, 11519–11524. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Tycko, R. Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Sci. 2010, 19, 716–730. [Google Scholar] [CrossRef] [Green Version]
- Bayro, M.J.; Chen, B.; Yau, W.-M.; Tycko, R. Site-Specific Structural Variations Accompanying Tubular Assembly of the HIV-1 Capsid Protein. J. Mol. Biol. 2014, 426, 1109–1127. [Google Scholar] [CrossRef] [Green Version]
- Bayro, M.J.; Ganser-Pornillos, B.K.; Zadrozny, K.K.; Yeager, M.; Tycko, R. Helical Conformation in the CA-SP1 Junction of the Immature HIV-1 Lattice Determined from Solid-State NMR of Virus-like Particles. J. Am. Chem. Soc. 2016, 138, 12029–12032. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef]
- Crowther, R.A.; Kiselev, N.A.; Böttcher, B.; Berriman, J.A.; Borisova, G.P.; Ose, V.; Pumpens, P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 1994, 77, 943–950. [Google Scholar] [CrossRef]
- Kenney, J.M.; von Bonsdorff, C.H.; Nassal, M.; Fuller, S.D. Evolutionary conservation in the hepatitis B virus core structure: Comparison of human and duck cores. Structure 1995, 3, 1009–1019. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Jin, L.; Jih, J.; Shih, C.; Hong Zhou, Z. 3.5 Å cryo-EM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein. PLoS ONE 2013, 8, e69729. [Google Scholar] [CrossRef] [Green Version]
- Nassal, M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 1992, 66, 4107–4116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summers, J.; Mason, W.S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell 1982, 29, 403–415. [Google Scholar] [CrossRef]
- Ning, X.; Nguyen, D.; Mentzer, L.; Adams, C.; Lee, H.; Ashley, R.; Hafenstein, S.; Hu, J. Secretion of Genome-Free Hepatitis B Virus—Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus. PLoS Pathog. 2011, 7, e1002255-14. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, L.; Wang, S.; Wiegand, T.; Bressanelli, S.; Nassal, M.; Meier, B.H.; Böckmann, A. Solid-state [13C-15N] NMR resonance assignment of hepatitis B virus core protein. Biomol. NMR Assign. 2018, 12, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Liu, F.; Tong, X.; Hoffmann, D.; Zuo, J.; Lu, M. Treatment of Chronic Hepatitis B Virus Infection Using Small Molecule Modulators of Nucleocapsid Assembly: Recent Advances and Perspectives. ACS Infect Dis. 2019, 5, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Verbinnen, T.; Tan, Y.; Wang, G.; Dehertogh, P.; Vergauwen, K.; Neefs, J.-M.; Jacoby, E.; Lenz, O.; Berke, J.M. Anti-HBV activity of the HBV capsid assembly modulator JNJ-56136379 across full-length genotype A–H clinical isolates and core site-directed mutants in vitro. J. Antimicrob. Chemother. 2020, 75, 2526–2534. [Google Scholar] [CrossRef]
- Schlicksup, C.J.; Laughlin, P.; Dunkelbarger, S.; Wang, J.C.-Y.; Zlotnick, A. Local Stabilization of Subunit–Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids. ACS Chem. Biol. 2020, 15, 1708–1717. [Google Scholar] [CrossRef]
- Barbet-Massin, E.; Felletti, M.; Schneider, R.; Jehle, S.; Communie, G.; Martinez, N.; Jensen, M.R.; Ruigrok, R.W.H.; Emsley, L.; Lesage, A.; et al. Insights into the structure and dynamics of measles virus nucleocapsids by 1H-detected solid-state NMR. Biophys. J. 2014, 107, 941–946. [Google Scholar] [CrossRef] [Green Version]
- Desfosses, A.; Goret, G.; Farias Estrozi, L.; Ruigrok, R.W.H.; Gutsche, I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 2011, 85, 1391–1395. [Google Scholar] [CrossRef] [Green Version]
- Gutsche, I.; Desfosses, A.; Effantin, G.; Ling, W.L.; Haupt, M.; Ruigrok, R.W.H.; Sachse, C.; Schoehn, G. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 2015, 348, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, B.S. Australia antigen and the biology of hepatitis B. Science 1977, 197, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heermann, K.H.; Goldmann, U.; Schwartz, W.; Seyffarth, T.; Baumgarten, H.; Gerlich, W.H. Large Surface Proteins of Hepatitis B Virus containing the Pre-s Sequence. J. Virol. 1984, 52, 396–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eble, B.E.; Lingappa, V.R.; Ganem, D. Hepatitis B surface antigen: An unusual secreted protein initially synthesized as a transmembrane polypeptide. Mol. Cell. Biol. 1986, 6, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Zahid, M.; Lünsdorf, H.; Rinas, U. Assessing stability and assembly of the hepatitis B surface antigen into virus-like particles during down-stream processing. Vaccine 2015, 33, 3739–3745. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J. Relating structure and function of viral membrane-spanning miniproteins. Curr. Opin. Virol. 2015, 12, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opella, S.J. Solid-state NMR and membrane proteins. J. Magn. Reson. 2015, 253, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Prorok, M.; Castellino, F.J.; Weliky, D.P. Oligomeric β-Structure of the Membrane-Bound HIV-1 Fusion Peptide Formed from Soluble Monomers. Biophys. J. 2004, 87, 1951–1963. [Google Scholar] [CrossRef] [Green Version]
- Wasniewski, C.M.; Parkanzky, P.D.; Bodner, M.L.; Weliky, D.P. Solid-state nuclear magnetic resonance studies of HIV and influenza fusion peptide orientations in membrane bilayers using stacked glass plate samples. Chem. Phys. Lipids 2004, 132, 89–100. [Google Scholar] [CrossRef]
- Bodner, M.L.; Gabrys, C.M.; Struppe, J.O.; Weliky, D.P. C13–C13 and N15–C13 correlation spectroscopy of membrane-associated and uniformly labeled human immunodeficiency virus and influenza fusion peptides: Amino acid-type assignments and evidence for multiple conformations. J. Chem. Phys. 2008, 128, 052319. [Google Scholar] [CrossRef]
- Yao, H.; Lee, M.W.; Waring, A.J.; Wong, G.C.L.; Hong, M. Viral fusion protein transmembrane domain adopts β-strand structure to facilitate membrane topological changes for virus–cell fusion. Proc. Natl. Acad. Sci. USA 2015, 112, 10926–10931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, E.P.; Curtis-Fisk, J.; Young, K.M.; Weliky, D.P. Solid-State Nuclear Magnetic Resonance (NMR) Spectroscopy of Human Immunodeficiency Virus gp41 Protein That Includes the Fusion Peptide: NMR Detection of Recombinant Fgp41 in Inclusion Bodies in Whole Bacterial Cells and Structural Characterization of Purified and Membrane-Associated Fgp41. Biochemistry 2011, 50, 10013–10026. [Google Scholar] [PubMed] [Green Version]
- Sackett, K.; Nethercott, M.J.; Zheng, Z.; Weliky, D.P. Solid-State NMR Spectroscopy of the HIV gp41 Membrane Fusion Protein Supports Intermolecular Antiparallel β Sheet Fusion Peptide Structure in the Final Six-Helix Bundle State. J. Mol. Biol. 2014, 426, 1077–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnayake, P.U.; Sackett, K.; Nethercott, M.J.; Weliky, D.P. pH-dependent vesicle fusion induced by the ectodomain of the human immunodeficiency virus membrane fusion protein gp41: Two kinetically distinct processes and fully-membrane-associated gp41 with predominant β sheet fusion peptide conformation. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Morgan, C.A.; Hong, M. Fully hydrophobic HIV gp41 adopts a hemifusion-like conformation in phospholipid bilayers. J. Biol. Chem. 2019, 294, 14732–14744. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, E.C.; Das, B.B.; Tian, Y.; Opella, S.J. Structural determination of virus protein U from HIV-1 by NMR in membrane environments. Biochim. Biophys. Acta 2015, 1848, 3007–3018. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; De Angelis, A.A.; Nevzorov, A.A.; Wu, C.H.; Opella, S.J. Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles. Biophys. J. 2006, 91, 3032–3042. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Mrse, A.A.; Nevzorov, A.A.; Mesleh, M.F.; Oblatt-Montal, M.; Montal, M.; Opella, S.J. Three-dimensional Structure of the Channel-forming Trans-membrane Domain of Virus Protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003, 333, 409–424. [Google Scholar] [CrossRef]
- Mandala, V.S.; Loftis, A.R.; Shcherbakov, A.A.; Pentelute, B.L.; Hong, M. Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism. Nat. Struct. Mol. Biol. 2020, 27, 160–167. [Google Scholar] [CrossRef]
- Lamb, R.A. The Structure, Function, and Pathobiology of the Influenza A and B Virus Ion Channels. Cold Spring Harbor Perspect. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Kim, S.; Kovacs, F.; Cross, T.A. Structure of the transmembrane region of the M2 protein H(+) channel. Protein Sci. 2001, 10, 2241–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, K.; Kim, S.; Zhang, L.; Cross, T.A. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry 2002, 41, 13170–13177. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Asbury, T.; Achuthan, S.; Li, C.; Bertram, R.; Quine, J.R.; Fu, R.; Cross, T.A. Backbone Structure of the Amantadine-Blocked Trans-Membrane Domain M2 Proton Channel from Influenza A Virus. Biophys. J. 2007, 92, 4335–4343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cady, S.D.; Mishanina, T.V.; Hong, M. Structure of Amantadine-Bound M2 Transmembrane Peptide of Influenza A in Lipid Bilayers from Magic-Angle-Spinning Solid-State NMR: The Role of Ser31 in Amantadine Binding. J. Mol. Biol. 2009, 385, 1127–1141. [Google Scholar] [CrossRef] [Green Version]
- Cady, S.D.; Schmidt-Rohr, K.; Wang, J.; Soto, C.S.; DeGrado, W.F.; Hong, M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Yi, M.; Dong, H.; Qin, H.; Peterson, E.; Busath, D.D.; Zhou, H.-X.; Cross, T.A. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 2010, 330, 509–512. [Google Scholar] [CrossRef] [Green Version]
- Andreas, L.B.; Reese, M.; Eddy, M.T.; Gelev, V.; Ni, Q.Z.; Miller, E.A.; Emsley, L.; Pintacuda, G.; Chou, J.J.; Griffin, R.G. Structure and Mechanism of the Influenza A M218-60 Dimer of Dimers. J. Am. Chem. Soc. 2015, 137, 14877–14886. [Google Scholar] [CrossRef] [Green Version]
- Stouffer, A.L.; Acharya, R.; Salom, D.; Levine, A.S.; Di Costanzo, L.; Soto, C.S.; Tereshko, V.; Nanda, V.; Stayrook, S.; DeGrado, W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451, 596–599. [Google Scholar] [CrossRef] [Green Version]
- Mandala, V.S.; Liao, S.Y.; Gelenter, M.D.; Hong, M. The Transmembrane Conformation of the Influenza B Virus M2 Protein in Lipid Bilayers. Sci. Rep. 2019, 9, 3725. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Hong, M. Investigation of the Curvature Induction and Membrane Localization of the Influenza Virus M2 Protein Using Static and Off-Magic-Angle Spinning Solid-State Nuclear Magnetic Resonance of Oriented Bicelles. Biochemistry 2015, 54, 2214–2226. [Google Scholar] [CrossRef] [Green Version]
- Cook, G.A.; Opella, S.J. NMR studies of p7 protein from hepatitis C virus. Eur. Biophys. J. 2010, 39, 1097–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, G.A.; Opella, S.J. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 1448–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, G.A.; Zhang, H.; Park, S.H.; Wang, Y.; Opella, S.J. Comparative NMR studies demonstrate profound differences between two viroporins: p7 of HCV and Vpu of HIV-1. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 554–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, G.A.; Dawson, L.A.; Tian, Y.; Opella, S.J. Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance. Biochemistry 2013, 52, 5295–5303. [Google Scholar] [CrossRef] [Green Version]
- Oestringer, B.P.; Bolivar, J.H.; Claridge, J.K.; Almanea, L.; Chipot, C.; Dehez, F.; Holzmann, N.; Schnell, J.R.; Zitzmann, N. Hepatitis C virus sequence divergence preserves p7 viroporin structural and dynamic features. Sci. Rep. 2019, 9, 8383. [Google Scholar] [CrossRef] [Green Version]
- Oestringer, B.P.; Bolivar, J.H.; Hensen, M.; Claridge, J.K.; Chipot, C.; Dehez, F.; Holzmann, N.; Zitzmann, N.; Schnell, J.R. Re-evaluating the p7 viroporin structure. Nature 2018, 562, E8–E18. [Google Scholar] [CrossRef]
- Chipot, C.; Dehez, F.; Schnell, J.R.; Zitzmann, N.; Pebay-Peyroula, E.; Catoire, L.J.; Miroux, B.; Kunji, E.R.S.; Veglia, G.; Cross, T.A.; et al. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem. Rev. 2018, 118, 3559–3607. [Google Scholar] [CrossRef] [Green Version]
- OuYang, B.; Xie, S.; Berardi, M.J.; Zhao, X.; Dev, J.; Yu, W.; Sun, B.; Chou, J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature 2013, 498, 521–525. [Google Scholar] [CrossRef]
- Gouttenoire, J.; Montserret, R.; Paul, D.; Castillo, R.; Meister, S.; Bartenschlager, R.; Penin, F.; Moradpour, D. Aminoterminal Amphipathic α-Helix AH1 of Hepatitis C Virus Nonstructural Protein 4B Possesses a Dual Role in RNA Replication and Virus Production. PLoS Pathog. 2014, 10, e1004501-17. [Google Scholar] [CrossRef]
- Moradpour, D.; Penin, F. Hepatitis C virus proteins: From structure to function. Curr. Top. Microbiol. Immunol. 2013, 369, 113–142. [Google Scholar]
- Briggs, E.L.A.; Gomes, R.G.B.; Elhussein, M.; Collier, W.; Findlow, I.S.; Khalid, S.; McCormick, C.J.; Williamson, P.T.F. Interaction between the NS4B amphipathic helix, AH2, and charged lipid headgroups alters membrane morphology and AH2 oligomeric state—Implications for the Hepatitis C virus life cycle. Biochim. Biophys. Acta (BBA) Biomembr. 2015, 1848, 1671–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogeron, M.-L.; Badillo, A.; Jirasko, V.; Gouttenoire, J.; Paul, D.; Lancien, L.; Moradpour, D.; Bartenschlager, R.; Meier, B.H.; Penin, F.; et al. Wheat germ cell-free expression: Two detergents with a low critical micelle concentration allow for production of soluble HCV membrane proteins. Protein Expr. Purif. 2015, 105, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Fogeron, M.-L.; Badillo, A.; Penin, F.; Böckmann, A. Wheat Germ Cell-Free Overexpression for the Production of Membrane Proteins. Methods Mol. Biol. 2017, 1635, 91–108. [Google Scholar] [PubMed]
- Fogeron, M.-L.; Paul, D.; Jirasko, V.; Montserret, R.; Lacabanne, D.; Molle, J.; Badillo, A.; Boukadida, C.; Georgeault, S.; Roingeard, P.; et al. Functional expression, purification, characterization, and membrane reconstitution of non-structural protein 2 from hepatitis C virus. Protein Expr. Purif. 2015, 116, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lacabanne, D.; Lends, A.; Danis, C.; Kunert, B.; Fogeron, M.-L.; Jirasko, V.; Chuilon, C.; Lecoq, L.; Orelle, C.; Chaptal, V.; et al. Gradient reconstitution of membrane proteins for solid-state NMR studies. J. Biomol. NMR 2017, 69, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Sborgi, L.; Ravotti, F.; Dandey, V.P.; Dick, M.S.; Mazur, A.; Reckel, S.; Chami, M.; Scherer, S.; Huber, M.; Böckmann, A.; et al. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 13237–13242. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Ferreira, R.; Taylor, N.M.; Arteni, A.-A.; Kumari, P.; Mona, D.; Ringler, P.; Britschgi, M.; Lauer, M.E.; Makky, A.; Verasdonck, J.; et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 2019, 8, e48907. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lecoq, L.; Fogeron, M.-L.; Meier, B.H.; Nassal, M.; Böckmann, A. Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies. Viruses 2020, 12, 1069. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101069
Lecoq L, Fogeron M-L, Meier BH, Nassal M, Böckmann A. Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies. Viruses. 2020; 12(10):1069. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101069
Chicago/Turabian StyleLecoq, Lauriane, Marie-Laure Fogeron, Beat H. Meier, Michael Nassal, and Anja Böckmann. 2020. "Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies" Viruses 12, no. 10: 1069. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101069






