Perspectives on Viral RNA Genomes and the RNA Folding Problem
Abstract
:1. Introduction
2. High Resolution Asymmetric Reconstructions Provide New Insights into Viral RNA Genome Structures
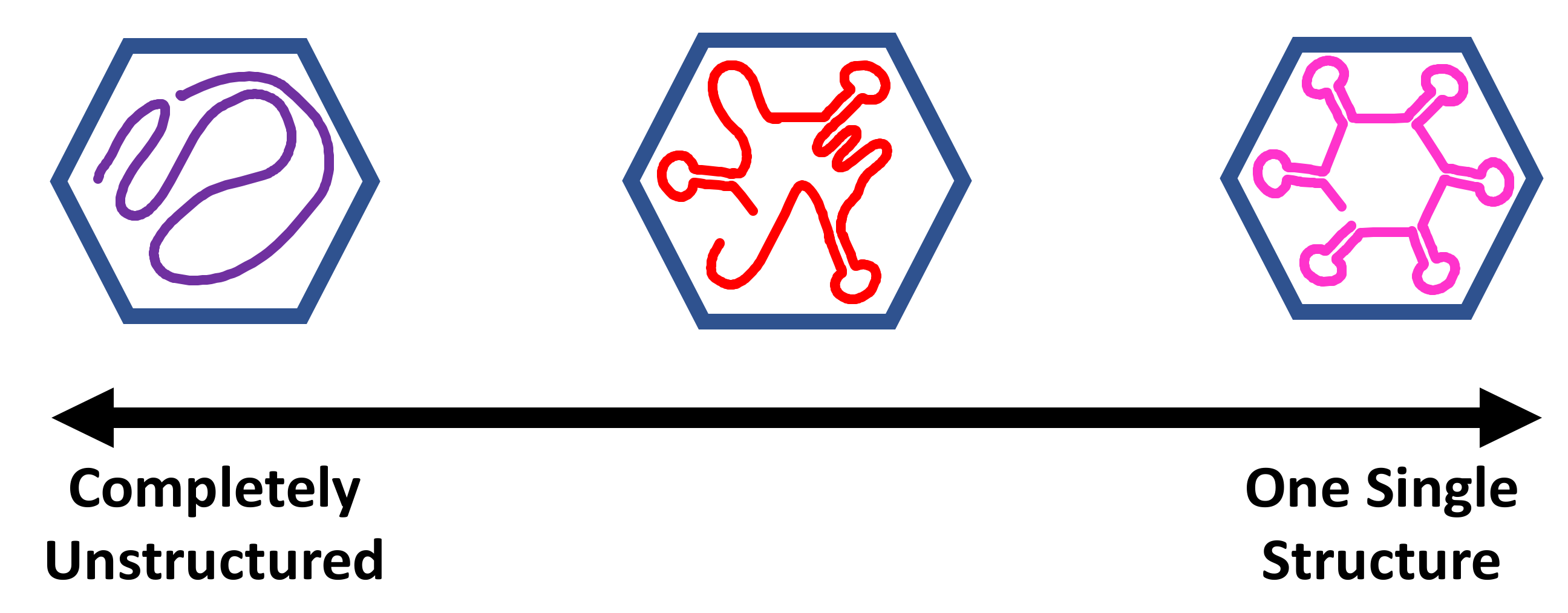
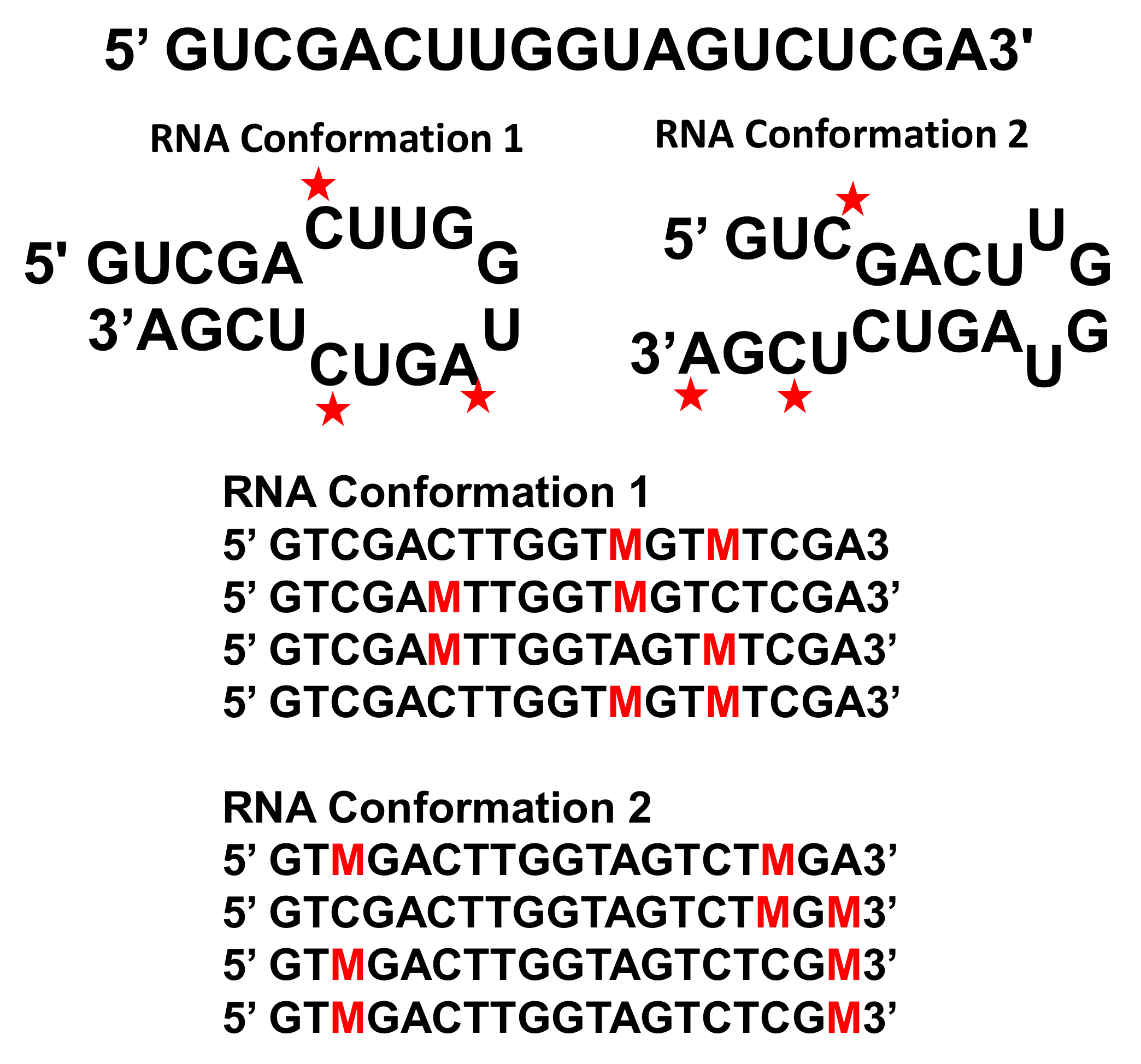
3. Advances in Chemical Probing Sequencing and Analysis Provide New Insights into Viral RNA Ensembles
4. Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, J.E.; Rossmann, M.G.; Smiley, I.E.; Wagner, M.A. Single crystal x-ray diffraction studies of Southern bean mosaic virus. J. Ultrastruct. Res. 1974, 46, 441–451. [Google Scholar] [CrossRef]
- Larson, S.B.; Koszelak, S.; Day, J.; Greenwood, A.; Dodds, J.A.; McPherson, A. Double Helical RNA in Stallite Tobacco Mosaic Virus. Nature 1993, 361, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.J.; Johnson, J.E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 1993, 361, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beren, C.; Cui, Y.; Chakravarty, A.; Yang, X.; Rao, A.L.N.; Knobler, C.M.; Zhou, Z.H.; Gelbart, W.M. Genome organization and interaction with capsid protein in a multipartite RNA virus. Proc. Natl. Acad. Sci. USA 2020, 117, 10673. [Google Scholar] [CrossRef]
- Tomezsko, P.J.; Corbin, V.D.A.; Gupta, P.; Swaminathan, H.; Glasgow, M.; Persad, S.; Edwards, M.D.; McIntosh, L.; Papenfuss, A.T.; Emery, A.; et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 2020, 582, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.-C.; Dettenhofer, M.; Yu, X.; Ehresmann, C.; Ehresmann, B.; Marquet, R. First Snapshots of the HIV-1 RNA Structure in Infected Cells and in Virions. J. Biol. Chem. 2004, 279, 48397–48403. [Google Scholar] [CrossRef] [Green Version]
- Sherpa, C.; Grice, S. Structural Fluidity of the Human Immunodeficiency Virus Rev Response Element. Viruses 2020, 12, 86. [Google Scholar] [CrossRef] [Green Version]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [Green Version]
- Villordo, S.M.; Filomatori, C.V.; Sánchez-Vargas, I.; Blair, C.D.; Gamarnik, A.V. Dengue Virus RNA Structure Specialization Facilitates Host Adaptation. PLoS Pathog. 2015. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, S.J.; Stone, J.W.; Bleckley, S.; Gibbons, T.; Mathews, D.M. Ensemble of Secondary Structures for Encapsidated Satellite Tobacco Mosaic Virus RNA Consistent with Chemical Probing and Crystallography Constraints. Biophys. J. 2011, 101, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, E.J.; Simpson, M.A.; Watts, N.J.; O’Kane, R.; Wang, B.; Erie, D.A.; McPherson, A.; Weeks, K.M. Long-range architecture in a viral RNA genome. Biochemistry 2013, 52, 3182–3190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Larson, S.B.; Heitsch, C.E.; McPherson, A.; Harvey, S.C. A model for the structure of satellite tobacco mosaic virus. J. Struct. Biol. 2012, 180, 110–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleckley, S.; Schroeder, S.J. Incorporating Global Features of RNA Motifs in Predictions for an Ensemble of Secondary Structures for Encapsidated MS2 Bacteriophage RNA. RNA 2012, 18, 1309–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolfsson, Ó.; Middleton, S.; Manfield, I.W.; White, S.J.; Fan, B.; Vaughan, R.; Ranson, N.A.; Dykeman, E.; Twarock, R.; Ford, J.; et al. Direct Evidence for Packaging Signal-Mediated Assembly of Bacteriophage MS2. J. Mol. Biol. 2016, 428, 431–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, S.J. Alternative Viewpoints and Alternative Structures for Satellite Tobacco Mosaic Virus RNA. Biochemistry 2014, 53, 6728–6737. [Google Scholar] [CrossRef]
- Patel, N.; Wroblewski, E.; Leonov, G.; Phillips, S.E.V.; Tuma, R.; Twarock, R.; Stockley, P.G. Rewriting nature’s assembly manual for a ssRNA virus. Proc. Natl. Acad. Sci. USA 2017, 114, 12255–12260. [Google Scholar] [CrossRef] [Green Version]
- Garmann, R.F.; Comas-Garcia, M.; Koay, M.S.T.; Cornelissen, J.J.L.M.; Knobler, C.M.; Gelbart, W.M. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J. Virol. 2014, 88, 10472–10479. [Google Scholar] [CrossRef] [Green Version]
- Borodavka, A.; Tuma, R.; Stockley, P.G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. USA 2012, 109, 15769. [Google Scholar] [CrossRef] [Green Version]
- Devkota, B.; Petrov, A.S.; Lemieux, S.; Boz, M.B.; Tang, L.; Schneemann, A.; Johnson, J.E.; Harvey, S.C. Structural and electrostatic characterization of pariacoto virus: Implications for viral assembly. Biopolymers 2009, 91, 530–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Routh, A. Mapping RNA-capsid interactions and RNA secondary structure within virus particles using next-generation sequencing. Nucl. Acids Res. 2020, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakeel, S.; Evans, J.D.; Hazelbaker, M.; Kao, C.C.; Vaughan, R.C.; Butcher, S.J. Intrinsically-disordered N-termini in human parechovirus 1 capsid proteins bind encapsidated RNA. Sci. Reports 2018, 8, 5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinothkumar, K.R.; Henderson, R. Single particle electron cryomicroscopy: Trends, issues and future perspective. Q. Rev. Biophys. 2016, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomezsko, P.; Swaminathan, H.; Rouskin, S. Viral RNA structure analysis using DMS-MaPseq. Methods 2020. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.J. Challenges and Approaches to Predicting RNA with Multiple Functional Structures. RNA 2018, 24, 1615–1624. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.Z.; Mathews, D.H. Experiment-Assisted Secondary Structure Prediction with RNA Structure. Methods Mol. Biol. 2016, 1490, 163–176. [Google Scholar]
- Lan, T.C.T.; Allan, M.F.; Malsick, L.E.; Khandwala, S.; Nyeo, S.S.Y.; Bathe, M.; Griffiths, A.; Rouskin, S. Structure of the full SARS-CoV-2 RNA genome in infected cells. bioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.06.29.178343v1 (accessed on 2 October 2020).
- Ishimaru, D.; Plant, E.P.; Sims, A.C.; Yount, B.L., Jr.; Roth, B.M.; Eldho, N.V.; Pérez-Alvarado, G.C.; Armbruster, D.W.; Baric, R.S.; Dinman, J.D.; et al. RNA dimerization plays a role in ribosomal frameshifting of the SARS coronavirus. Nucl. Acids Res. 2013, 41, 2594–2608. [Google Scholar] [CrossRef]
- Kelly, J.A.; Olson, A.N.; Neupane, K.; Munshi, S.; San Emeterio, J.; Pollack, L.; Woodside, M.T.; Dinman, J.D. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 2020, 295, 10741–10748. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schroeder, S.J. Perspectives on Viral RNA Genomes and the RNA Folding Problem. Viruses 2020, 12, 1126. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101126
Schroeder SJ. Perspectives on Viral RNA Genomes and the RNA Folding Problem. Viruses. 2020; 12(10):1126. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101126
Chicago/Turabian StyleSchroeder, Susan J. 2020. "Perspectives on Viral RNA Genomes and the RNA Folding Problem" Viruses 12, no. 10: 1126. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101126



