Frequency and Duration of SARS-CoV-2 Shedding in Oral Fluid Samples Assessed by a Modified Commercial Rapid Molecular Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Specimens Collection
2.2. Analytic Sensitivity
2.3. Analytic Specificity
2.4. Simplexa™ COVID-19 Direct Assay
2.5. Statistical Analysis
2.6. Ethical statement
3. Results
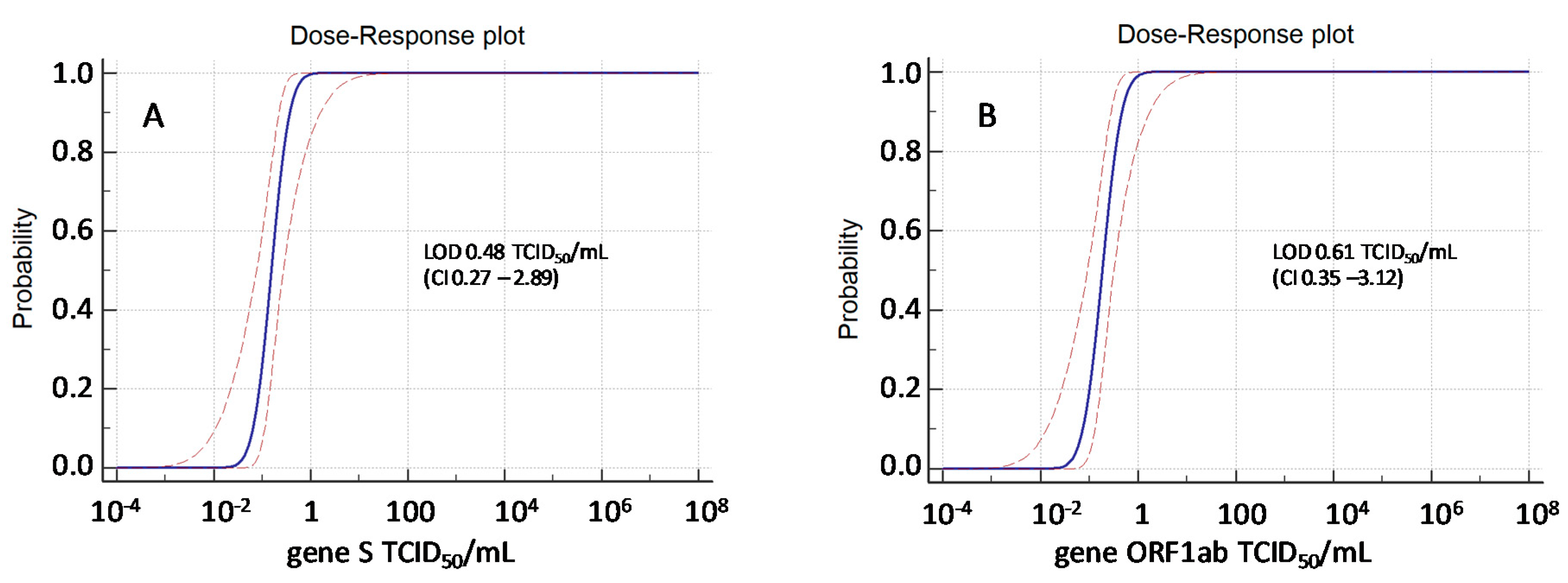
3.1. Analytical Sensitivity and Lower Limit of Detection (LOD)
3.2. Analytic Specificity
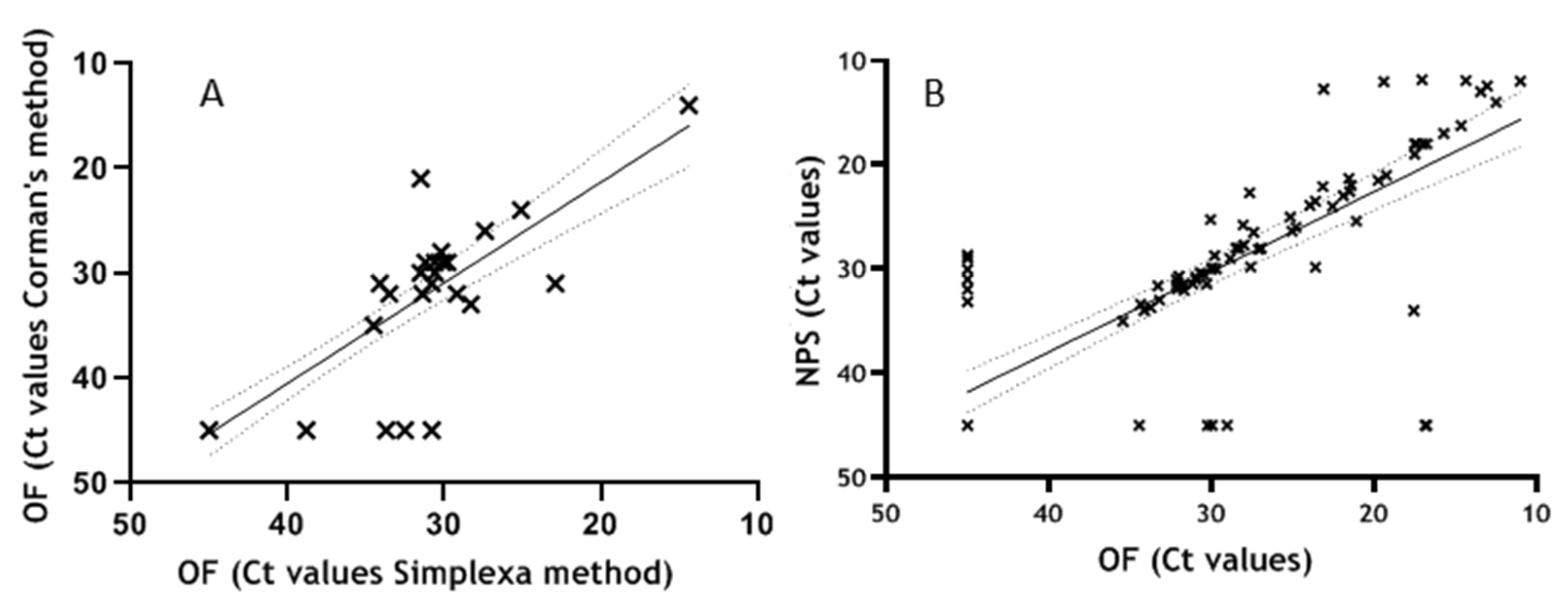
3.3. Performance Evaluation on Clinical Specimens
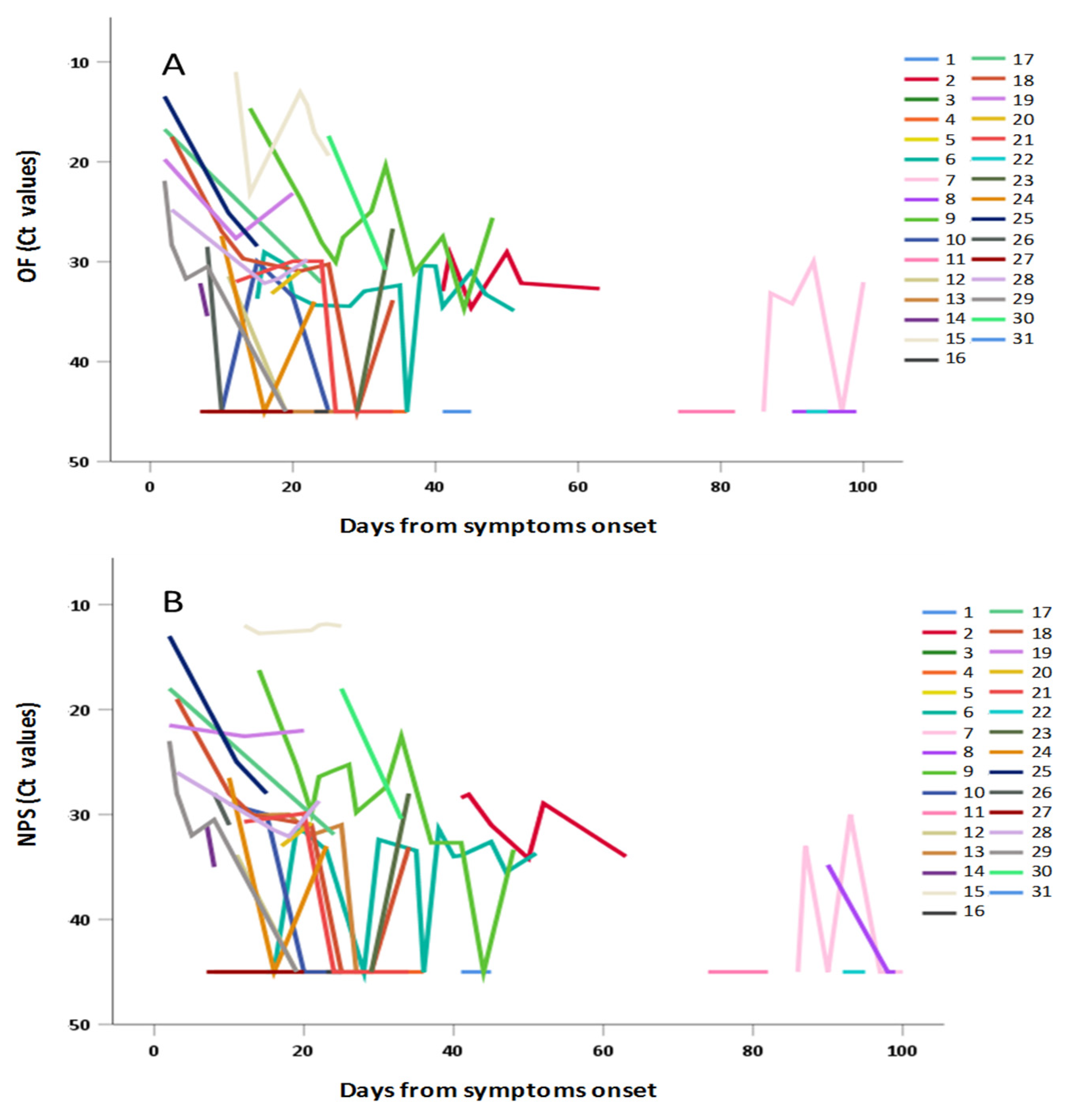
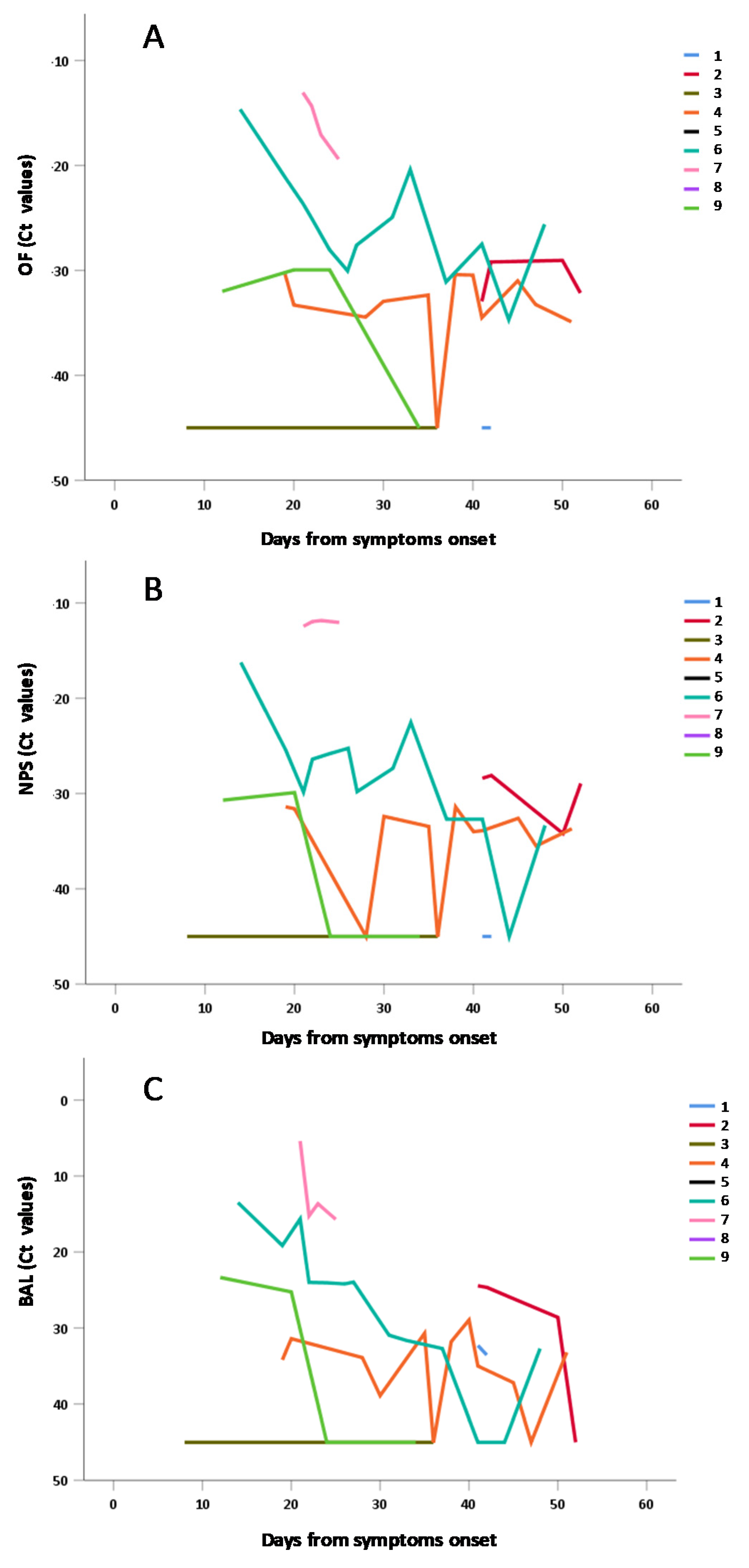
3.4. Frequency and Duration of SARS-CoV-2 Shedding
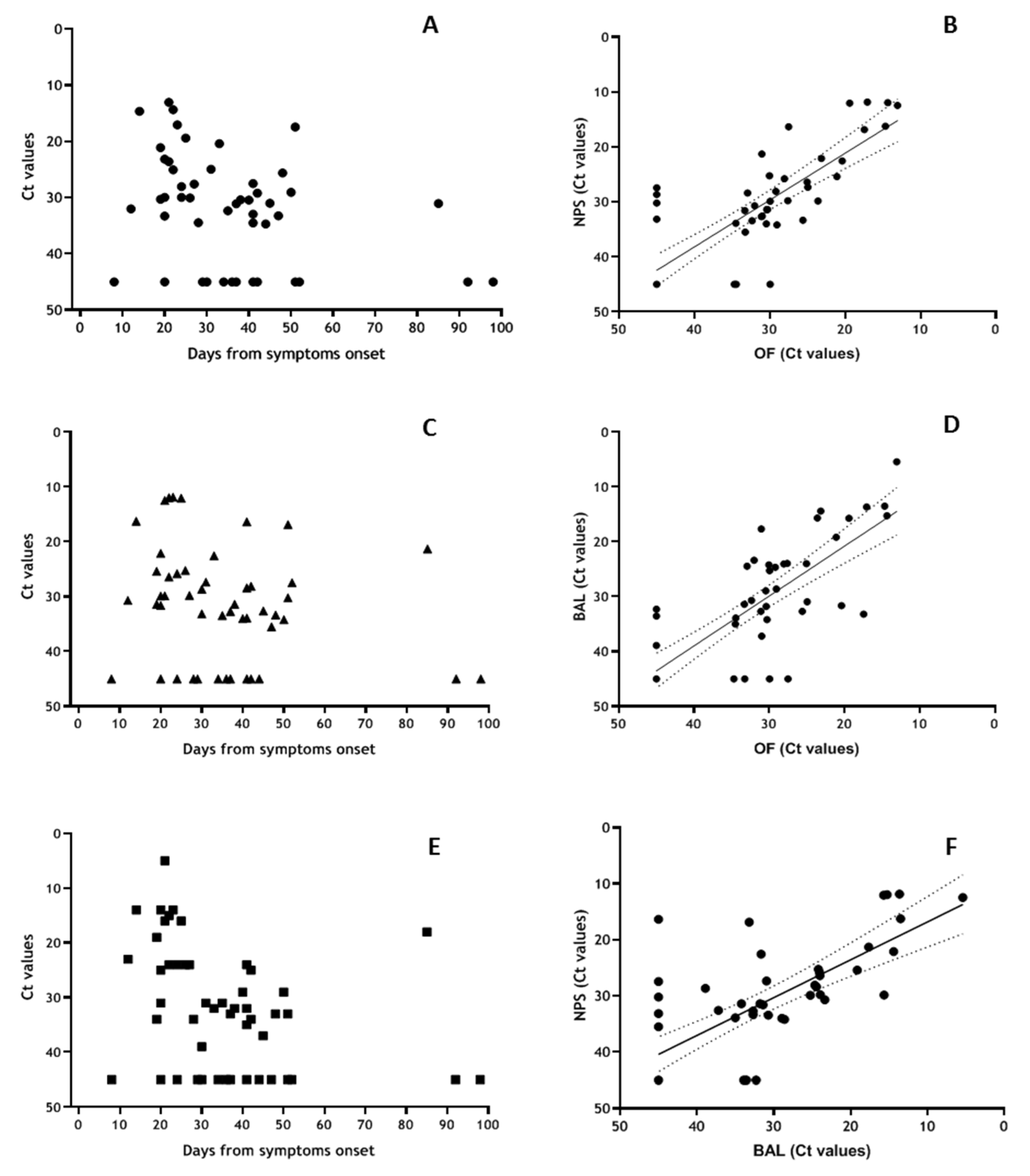
3.5. Sub-Group Analyses for OF vs. NPS vs. BAL Samples
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef] [PubMed]
- The World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases. Interim guidance 2 March 2020. Available online: https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 (accessed on 18 March 2020).
- ECDC. Novel Coronavirus (SARS-CoV-2) Discharge Criteria for Confirmed COVID-19 Cases e when Is It Safe to Discharge COVID-19 Cases from the Hospital or end Home Isolation? Available online: https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-Discharge-criteria.pdf (accessed on 29 March 2020).
- Zhu, J.; Guo, J.; Xu, Y.; Chen, X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J. Infect. 2020, 81, e48–e50. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Contreras, J.; Espinoza, M.A.; Sandoval-Jaime, C.; Cantú-Cuevas, M.A.; Barón-Olivares, H.; Ortiz-Orozco, O.D.; Muñoz-Rangel, A.V.; La Cruz, M.H.-D.; Eroza-Osorio, C.M.; Arias, C.F.; et al. Saliva sampling and its direct lysis, an excellent option to increase the number of SARS CoV2 diagnostic tests in settings with supply shortages. J. Clin. Microbiol. 2020, 58, 01659-20. [Google Scholar] [CrossRef]
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; A Williamson, D. Saliva as a Noninvasive Specimen for Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription–Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, 01438-20. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Fujisawa, S.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Fukumoto, T.; Sato, K.; Oguri, S.; Taki, K.; Senjo, H.; et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020, 81, e145–e147. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- Pasomsub, E.; Watcharananan, S.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: A cross-sectional study. Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hamid, H.; Khurshid, Z.; Adanir, N.; Zafar, M.S.; Zohaib, S. COVID-19 Pandemic and Role of Human Saliva as a Testing Biofluid in Point-of-Care Technology. Eur. J. Dent. 2020. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Tajima, Y.; Suda, Y.; Yano, K. A case report of SARS-CoV-2 confirmed in saliva specimens up to 37 days after onset: Proposal of saliva specimens for COVID-19 diagnosis and virus monitoring. J. Infect. Chemother. 2020, 26, 1086–1089. [Google Scholar] [CrossRef] [PubMed]
- Capobianchi, M.R.; Rueca, M.; Messina, F.; Giombini, E.; Carletti, F.; Colavita, F.; Castilletti, C.; Lalle, E.; Bordi, L.; Vairo, F.; et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin. Microbiol. Infect. 2020, 26, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef] [Green Version]
- Bordi, L.; Piralla, A.; Lalle, E.; Giardina, F.; Colavita, F.; Tallarita, M.; Sberna, G.; Novazzi, F.; Meschi, S.; Castilletti, C.; et al. Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa™ COVID-19 direct assay. J. Clin. Virol. 2020, 128, 104416. [Google Scholar] [CrossRef] [PubMed]
- Uhteg, K.; Jarrett, J.; Richards, M.; Howard, C.; Morehead, E.; Geahr, M.; Gluck, L.; Hanlon, A.; Ellis, B.; Kaur, H.; et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020, 127, 104384. [Google Scholar] [CrossRef] [PubMed]
- Czumbel, L.M.; Kiss, S.; Farkas, N.; Mandel, I.; Hegyi, A.; Nagy, Á.; Lohinai, Z.; Szakács, Z.; Hegyi, P.; Steward, M.C.; et al. Saliva as a Candidate for COVID-19 Diagnostic Testing: A Meta-Analysis. Front. Med. 2020, 7, 465. [Google Scholar] [CrossRef] [PubMed]

| Asymptomatic | Paucisymptomatic | Severe (PaO2/FiO2 < 100) | Negative | Tot | |
|---|---|---|---|---|---|
| Patients (N°) | 14 | 61 | 12 | 77 | 164 |
| Samples (N°) | 18 | 154 | 50 | 115 | 337 |
| OF: Median Ct Values (Range) | NPS: Median Ct Values (Range) | ||
|---|---|---|---|
| Tot | 32.2 (11.0–45) | 32.0 (11.9–45) | |
| DSO | 0–30 | 31.0 (11.0–45) | 31.0 (11.9–45) |
| 31–60 | 33.3 (20.4–45) | 33.4 (22.6–45) | |
| >60 | 45.0 (30.0–45) | 45.0 (21.3–45) |
| Age | Significance (p Value) | ||
|---|---|---|---|
| <60 Years | >60 Years | ||
| OF: Median Ct values (range) | 29.9 (13.5–45) | 34.3 (11.0–45) | p = 0.0007 |
| NPS: Median Ct values (range) | 29.8 (13.0–45) | 33.9 (11.9–45) | p = 0.0004 |
| OF: Median Ct Values (Range) | NPS: Median Ct Values (Range) | BAL: Median Ct Values (Range) | ||
|---|---|---|---|---|
| Tot | 31.0 (13.1–45) | 31.4 (5–45) | 32.5 (11.9–45) | |
| DSO | 0–30 | 30.0 (13.1–45) | 29.8 (11.9–45) | 24.1 (5.0–45) |
| 31–60 | 32.7 (17.5–45) | 33.4 (16.4–45) | 33.4 (24.5–45) | |
| >60 | 45 (31.1–45) | 45 (21.3–45) | 45 (17.7–45) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordi, L.; Sberna, G.; Lalle, E.; Piselli, P.; Colavita, F.; Nicastri, E.; Antinori, A.; Boumis, E.; Petrosillo, N.; Marchioni, L.; et al. Frequency and Duration of SARS-CoV-2 Shedding in Oral Fluid Samples Assessed by a Modified Commercial Rapid Molecular Assay. Viruses 2020, 12, 1184. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101184
Bordi L, Sberna G, Lalle E, Piselli P, Colavita F, Nicastri E, Antinori A, Boumis E, Petrosillo N, Marchioni L, et al. Frequency and Duration of SARS-CoV-2 Shedding in Oral Fluid Samples Assessed by a Modified Commercial Rapid Molecular Assay. Viruses. 2020; 12(10):1184. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101184
Chicago/Turabian StyleBordi, Licia, Giuseppe Sberna, Eleonora Lalle, Pierluca Piselli, Francesca Colavita, Emanuele Nicastri, Andrea Antinori, Evangelo Boumis, Nicola Petrosillo, Luisa Marchioni, and et al. 2020. "Frequency and Duration of SARS-CoV-2 Shedding in Oral Fluid Samples Assessed by a Modified Commercial Rapid Molecular Assay" Viruses 12, no. 10: 1184. https://0-doi-org.brum.beds.ac.uk/10.3390/v12101184





