Manipulation of Jasmonate Signaling by Plant Viruses and Their Insect Vectors
Abstract
:1. Introduction
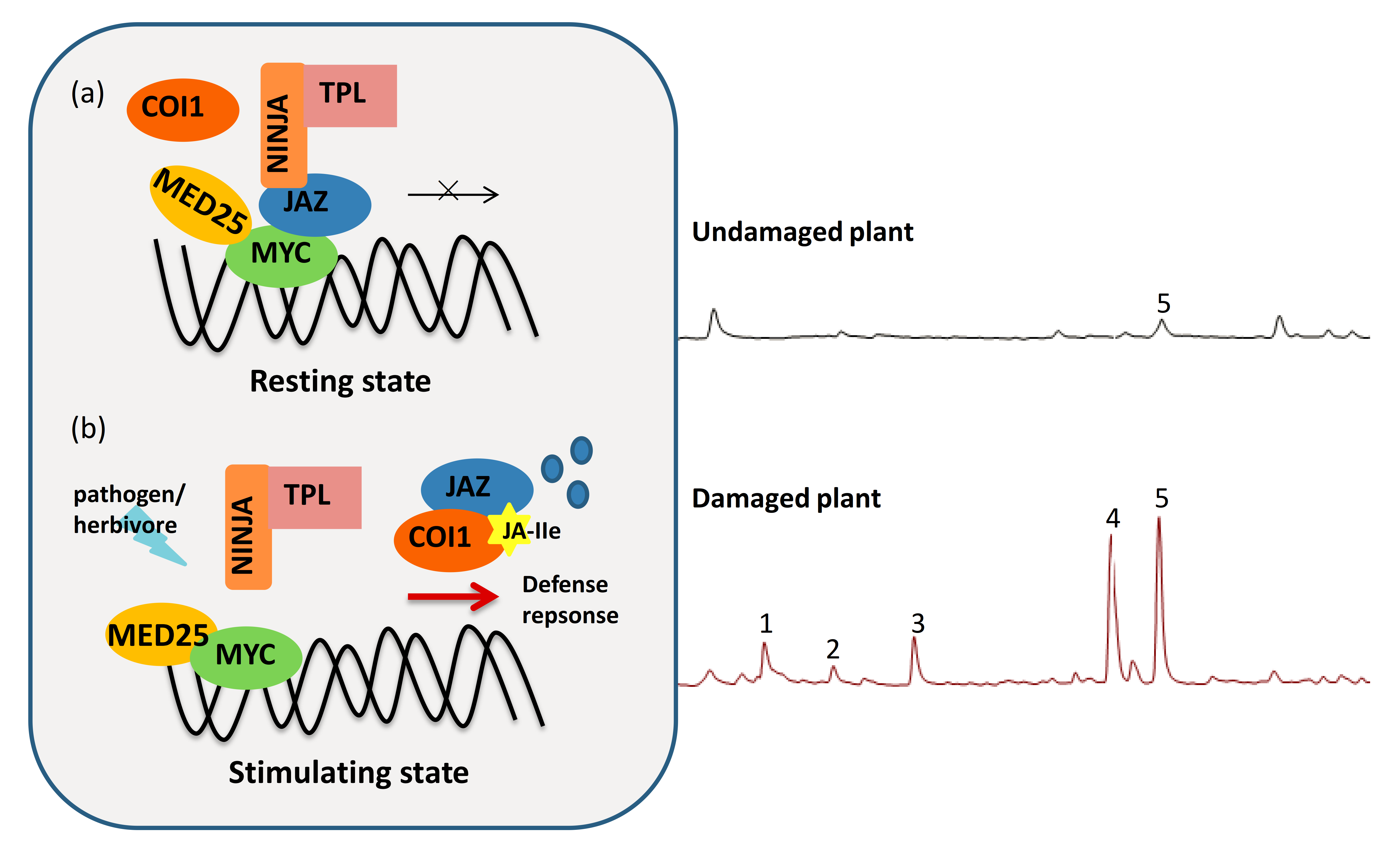
2. Overview of Jasmonate Derivatives (JAs) and JA Signaling
3. The Counter-Defense of Virus and Herbivore to JA Signaling in Plants
3.1. The Herbivore Manipulation of JA Signaling
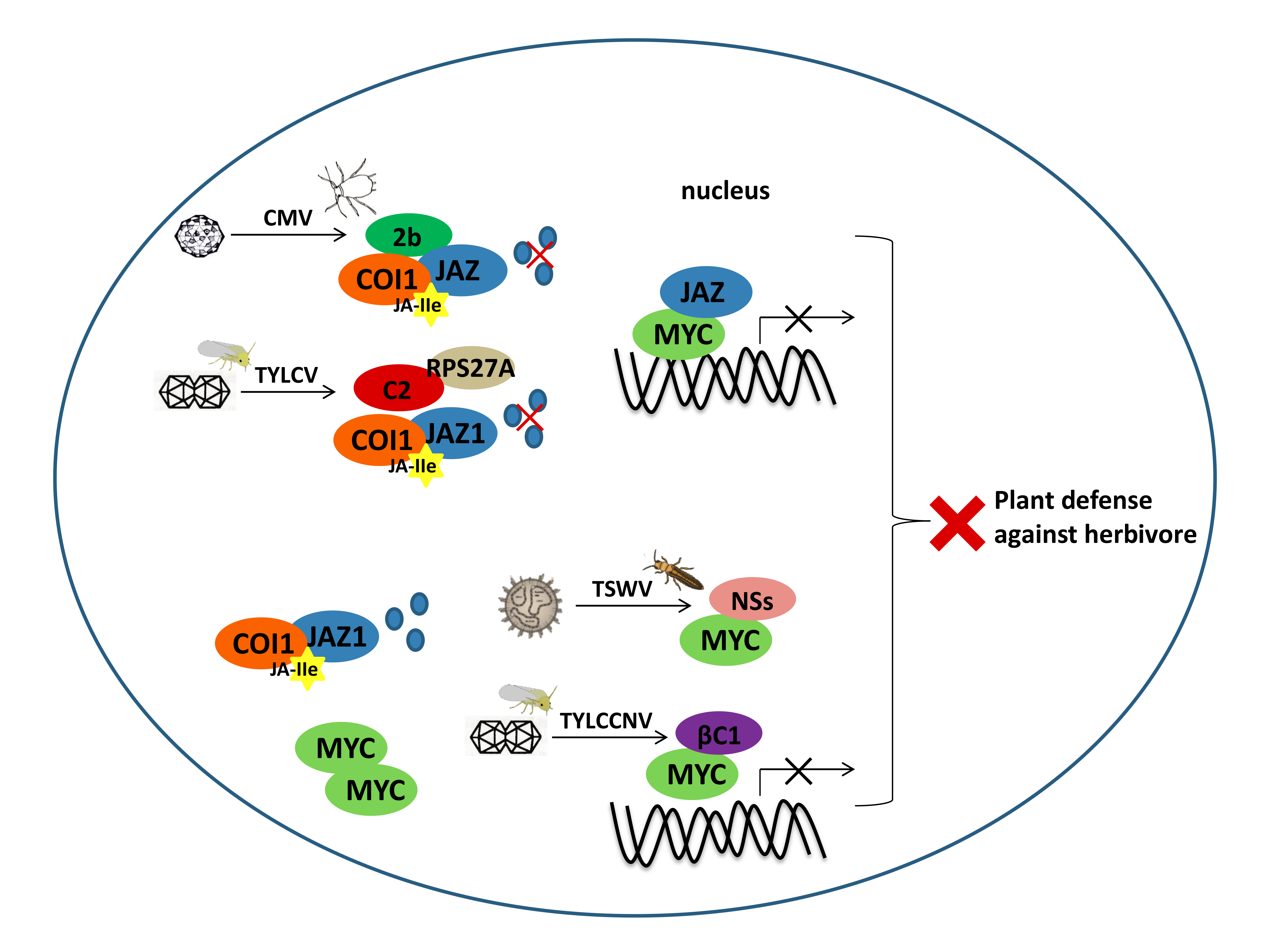
3.2. JAZ-MYC is one of the Common Targets of Plant Viruses
3.3. JA-Regulated Chemical Defense Hijacked by Plant Viruses
4. The Plant Defense-Growth Trade-Off Regulated by JAs
5. Perspectives
Funding
Conflicts of Interest
References
- Kennedy, S.J. Benefits to Aphids from Feeding on Galled and Virus-infected Leaves. Nature 1951, 168, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrode, S.D.; Bosque-Pérez, N.; Davis, T.S. Insect-Borne Plant Pathogens and Their Vectors: Ecology, Evolution, and Complex Interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.L. Effect of Tomato Mottle Virus (ToMoV) on Bemisia tabaci Biotype B (Homoptera: Aleyrodidae) Oviposition and Adult Survivorship on Healthy Tomato. Fla. Entomol. 2002. [Google Scholar] [CrossRef]
- Shrestha, A.; Srinivasan, R.; Riley, D.G.; Culreath, A. Direct and indirect effects of a thrips-transmitted Tospovirus on the preference and fitness of its vector, Frankliniella fusca. Entomol. Exp. Appl. 2012, 145, 260–271. [Google Scholar] [CrossRef]
- Czosnek, H.; Rubinstein, G. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 1997, 78, 2683–2689. [Google Scholar]
- Saioa, L.; Apurba, B.; Wendy, M.; Stan, D.; Rajagopalbabu, S.; Youjun, Z. Temporal Effects of a Begomovirus Infection and Host Plant Resistance on the Preference and Development of an Insect Vector, Bemisia tabaci, and Implications for Epidemics. PLoS ONE 2015, 10, e0142114. [Google Scholar]
- Abe, H.; Tomitaka, Y.; Shimoda, T.; Seo, S.; Sakurai, T.; Kugimiya, S.; Tsuda, S.; Kobayashi, M. Antagonistic Plant Defense System Regulated by Phytohormones Assists Interactions Among Vector Insect, Thrips and a Tospovirus. Plant Cell Physiol. 2012, 53, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Medina-Ortega, K.J.; Bosque-Pérez, N.A.; Ngumbi, E.; Jiménez-Martínez, E.S.; Eigenbrode, S.D. Rhopalosiphum padi (Hemiptera: Aphididae) Responses to Volatile Cues From Barley Yellow Dwarf Virus–Infected Wheat. Environ. Entomol. 2009, 38, 836–845. [Google Scholar] [CrossRef] [Green Version]
- Eigenbrode, S.D.; Ding, H.; Shiel, P.; Berger, P.H. Volatiles from potato plants infected with potato leafroll virus attract and arrest the virus vector, Myzus persicae (Homoptera: Aphididae). Proc. Biol. Sci. 2002, 269, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Carmo-Sousa, M.; Moreno, A.; Garzo, E.; Fereres, A. A non-persistently transmitted-virus induces a pull-push strategy in its aphid vector to optimize transmission and spread. Virus Res. 2014, 186, 38–46. [Google Scholar] [CrossRef]
- Carmo-Sousa, M.; Moreno, A.; Plaza, M.; Garzo, E.; Fereres, A. Cucurbit aphid-borne yellows virus (CABYV) modifies the alighting, settling and probing behaviour of its vector Aphis gossypii favouring its own spread. Ann. Appl. Biol. 2016, 169, 284–297. [Google Scholar] [CrossRef]
- Casteel, C.L.; De Alwis, M.; Bak, A.; Dong, H.; Whitham, S.A.; Jander, G. Disruption of Ethylene Responses by Turnip mosaic virus Mediates Suppression of Plant Defense against the Green Peach Aphid Vector. Plant Physiol. 2015, 169, 209–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casteel, C.L.; Yang, C.; Nanduri, A.C.; Jong, H.N.D.; Jander, G. The NIa-Pro Protein of Turnip mosaic virus Improves Growth and Reproduction of the Aphid Vector, Myzus persicae (Green Peach Aphid). Plant J. 2013, 77, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Groen, S.C.; Du, Z.; Murphy, A.M.; Anggoro, D.T.; Tungadi, T.; Luang-In, V.; Lewsey, M.G.; Rossiter, J.T.; Powell, G.; et al. A trio of viral proteins tunes aphid-plant interactions in Arabidopsis thaliana. PLoS ONE 2013, 8, e83066. [Google Scholar] [CrossRef] [Green Version]
- Shalileh, S.; Ogada, P.A.; Moualeu, D.P.; Poehling, H.-M. Manipulation of Frankliniella occidentalis (Thysanoptera: Thripidae) by Tomato Spotted Wilt Virus (Tospovirus) Via the Host Plant Nutrients to Enhance Its Transmission and Spread. Environ. Entomol. 2016, 45, 1235–1242. [Google Scholar] [CrossRef] [Green Version]
- Bosque-Pérez, N.A.; Eigenbrode, S.D. The influence of virus-induced changes in plants on aphid vectors: Insights from luteovirus pathosystems. Virus Res. 2011, 159, 201–205. [Google Scholar] [CrossRef]
- Lortzing, T.; Steppuhn, A. Jasmonate signalling in plants shapes plant–insect interaction ecology. Curr. Opin. Insect Sci. 2016, 14, 32–39. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The Role of the Jasmonate Response in Plant Susceptibility to Diverse Pathogens with a Range of Lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Tee, C.S.; Jiang, Y.L.; Jiang, X.Y.; Venkatesh, P.N.; Sarojam, R.; Ye, J. A terpenoid phytoalexin plays a role in basal defense of Nicotiana benthamiana against Potato virus X. Sci. Rep. 2015, 5, 9682. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Marcos, A.; Pacheco, R.; Manzano, A.; Aguilar, E.; Tenllado, F. Oxylipin Biosynthesis Genes Positively Regulate Programmed Cell Death during Compatible Infections with the Synergistic Pair Potato Virus X-Potato Virus Y and Tomato Spotted Wilt Virus. J. Virol. 2013, 87, 5769–5783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, R.; Garcia-Marcos, A.; Manzano, A.; de Lacoba, M.G.; Camanes, G.; Garcia-Agustin, P.; Diaz-Ruiz, J.R.; Tenllado, F. Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant-virus interactions that lead to cell death. Mol. Plant Microbe Interact. 2012, 25, 709–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozanodurán, R.; Rosasdíaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Xing, W.D.; Bejarano, E.R. Geminiviruses Subvert Ubiquitination by Altering CSN-Mediated Derubylation of SCF E3 Ligase Complexes and Inhibit Jasmonate Signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 2017, 37, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Ahammed, G.; Wu, C.; Fan, S.y.; Zhou, Y.H. Crosstalk among Jasmonate, Salicylate and Ethylene Signaling Pathways in Plant Disease and Immune Responses. Curr. Protein Pept. Sci. 2015, 16, 450–461. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Shi, J.H.; Sun, Z.; Hu, X.J.; Jin, H.; Foba, C.N.; Liu, H.; Wang, C.; Liu, L.; Li, F.F.; Wang, M.Q. Rice defense responses are induced upon leaf rolling by an insect herbivore. BMC Plant Biol. 2019, 19, 514. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Ibarra-Laclette, E.; Adame-Alvarez, R.M.; Martinez, O.; Ramirez-Chavez, E.; Molina-Torres, J.; Herrera-Estrella, L. How plants sense wounds: Damaged-self recognition is based on plant-derived elicitors and induces octadecanoid signaling. PLoS ONE 2012, 7, e30537. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, F.; Bodenhausen, N.; Lassueur, S.; Masclaux, F.G.; Reymond, P. Differential Contribution of Transcription Factors to Arabidopsis thaliana Defense Against Spodoptera littoralis. Front. Plant Sci. 2013, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, K.; Meng, Y.; Hu, J.; Ding, M.; Bian, J.; Yan, M.; Han, J.; Zhou, M. Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA59 transcription factor. Plant J. 2018, 95, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Engelberth, J.; Alborn, H.T.; Schmelz, E.A.; Tumlinson, J.H. Airborne signals prime plants against insect herbivore attack. Proc. Natl. Acad. Sci. USA 2004, 101, 1781–1785. [Google Scholar] [CrossRef] [Green Version]
- Ton, J.; D’Alessandro, M.; Jourdie, V.; Jakab, G.; Karlen, D.; Held, M.; Mauch-Mani, B.; Turlings, T.C. Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 2007, 49, 16–26. [Google Scholar] [CrossRef] [Green Version]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Metraux, J.P.; Van Loon, L.C.; Dicke, M.; et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef]
- Wu, J.; Hettenhausen, C.; Meldau, S.; Baldwin, I.T. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 2007, 19, 1096–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wang, D.D.; Fang, X.; Chen, X.Y.; Mao, Y.B. Plant Specialized Metabolism Regulated by Jasmonate Signaling. Plant Cell Physiol. 2019, 60, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2017, 214, 388–399. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB betaC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef]
- Joo, Y.; Schuman, M.C.; Goldberg, J.K.; Wissgott, A.; Kim, S.G.; Baldwin, I.T. Herbivory elicits changes in green leaf volatile production via jasmonate signaling and the circadian clock. Plant Cell Environ. 2019, 42, 972–982. [Google Scholar] [CrossRef]
- Schmiesing, A.; Emonet, A. Arabidopsis MYC Transcription Factors Are the Target of Hormonal Salicylic Acid/Jasmonic Acid Cross Talk in Response to Pieris brassicae Egg Extract. Plant Physiol. 2016, 170, 2432–2443. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.J.; Huang, F.; Zhang, J.M.; Wei, J.N.; Lu, Y.B. The mealybug Phenacoccus solenopsis suppresses plant defense responses by manipulating JA-SA crosstalk. Sci. Rep. 2015, 5, 9354. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.I.; Prince, D.; Pitino, M.; Maffei, M.E.; Win, J.; Hogenhout, S.A. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 2010, 6, e1001216. [Google Scholar] [CrossRef]
- Will, T.; Tjallingii, W.F.; Thonnessen, A.; van Bel, A.J. Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. USA 2007, 104, 10536–10541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutti, N.S.; Louis, J.; Pappan, L.K.; Pappan, K.; Begum, K.; Chen, M.S.; Park, Y.; Dittmer, N.; Marshall, J.; Reese, J.C.; et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA 2008, 105, 9965–9969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, R.; Ye, W.; Chen, H.; Zeng, J. A Salivary Endo-beta-1,4-Glucanase Acts as an Effector That Enables the Brown Planthopper to Feed on Rice. Plant Physiol. 2017, 173, 1920–1932. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Escudero-Martinez, C. An Aphid Effector Targets Trafficking Protein VPS52 in a Host-Specific Manner to Promote Virulence. Plant Physiol. 2017, 173, 1892–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Khan, A.N.; Subrahmanyam, S.; Raman, A.; Taylor, G.S.; Fletcher, M.J. Salivary proteins of plant-feeding hemipteroids - implication in phytophagy. Bull. Entomol. Res. 2014, 104, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.X.; Qian, L.X.; Wang, X.W.; Shao, R.X.; Hong, Y.; Liu, S.S.; Wang, X.W. A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Diezel, C.; von Dahl, C.C.; Gaquerel, E.; Baldwin, I.T. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol. 2009, 150, 1576–1586. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Zhao, P.; Ma, Y.; Yao, X.; Sun, Y.; Huang, X.; Jin, J.; Zhang, Y.; Zhu, C.; Fang, R.; et al. A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos. Trans. R Soc. Lond. B Biol. Sci. 2019, 374, 20180313. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Y.; Liu, Y.Q.; Song, W.M.; Chen, D.Y.; Chen, F.Y.; Chen, X.Y.; Chen, Z.W.; Ge, S.X.; Wang, C.Z. An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14331–14338. [Google Scholar] [CrossRef] [Green Version]
- Musser, R.O.; Hum-Musser, S.M.; Eichenseer, H.; Peiffer, M.; Ervin, G.; Murphy, J.B.; Felton, G.W. Herbivory: Caterpillar saliva beats plant defences. Nature 2002, 416, 599–600. [Google Scholar] [CrossRef] [PubMed]
- Sonderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Malka, O.; Shekhov, A.; Reichelt, M.; Gershenzon, J.; Vassao, D.G.; Morin, S. Glucosinolate Desulfation by the Phloem-Feeding Insect Bemisia tabaci. J. Chem. Ecol. 2016, 42, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Lewsey, M.G.; Murphy, A.M.; Maclean, D.; Dalchau, N.; Westwood, J.H.; Macaulay, K.; Bennett, M.H.; Moulin, M.; Hanke, D.E.; Powell, G.; et al. Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol. Plant Microbe Interact. 2010, 23, 835–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csorba, T.; Kontra, L.; Burgyan, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhao, J. MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Liu, C.; Deng, W.H.; Yao, D.M.; Pan, L.L.; Li, Y.Q.; Liu, Y.Q.; Liang, Y.; Zhou, X.P.; Wang, X.W. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 2019, 15, e1007607. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Xu, S.; Zhao, P. The Orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog. 2019, 15, e1007897. [Google Scholar] [CrossRef]
- Wu, D.; Qi, T.; Li, W.X.; Tian, H.; Gao, H.; Wang, J.; Ge, J.; Yao, R.; Ren, C.; Wang, X.B.; et al. Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 2017, 27, 402–415. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Boter, M.; Fernandez-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Yao, J.; Ma, K.W.; Zhou, H.; Song, J.; He, S.Y.; Ma, W. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssieres, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Teixeira, P.J.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas syringae Type III Effector HopBB1 Promotes Host Transcriptional Repressor Degradation to Regulate Phytohormone Responses and Virulence. Cell Host Microbe. 2017, 21, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Monte, I.; Franco-Zorrilla, J.M.; Garcia-Casado, G.; Zamarreno, A.M.; Garcia-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant. 2019, 12, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Bigotes, A.; Valenzuela-Riffo, F.; Figueroa, C.R. Evolutionary Analysis of JAZ Proteins in Plants: An Approach in Search of the Ancestral Sequence. Int. J. Mol. Sci. 2019, 20, 5060. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Su, Q.; Shi, X.; Pan, H.; Jiao, X.; Zhang, Y. Persistently Transmitted Viruses Restrict the Transmission of Other Viruses by Affecting Their Vectors. Front. Physiol. 2018, 9, 1261. [Google Scholar] [CrossRef]
- Yang, J.Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.H. betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Yao, X.; Cai, C.; Li, R.; Du, J.; Sun, Y.; Wang, M.; Zou, Z.; Wang, Q.; Kliebenstein, D.J. Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci. Adv. 2019, 5, eaav9801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Kugler, A.; McGale, E.; Haverkamp, A.; Knaden, M.; Guo, H.; Beran, F.; Yon, F.; Li, R.; Lackus, N.; et al. Tissue-Specific Emission of (E)-alpha-Bergamotene Helps Resolve the Dilemma When Pollinators Are Also Herbivores. Curr. Biol. 2017, 27, 1336–1341. [Google Scholar] [CrossRef] [Green Version]
- Alquezar, B.; Volpe, H.X.L.; Magnani, R.F.; de Miranda, M.P.; Santos, M.A.; Wulff, N.A.; Bento, J.M.S.; Parra, J.R.P.; Bouwmeester, H.; Pena, L. beta-caryophyllene emitted from a transgenic Arabidopsis or chemical dispenser repels Diaphorina citri, vector of Candidatus Liberibacters. Sci. Rep. 2017, 7, 5639. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, G.; Huang, X.; Guo, H.; Su, X.; Han, L.; Zhang, Y.; Qi, Z.; Xiao, Y. Overexpression of the caryophyllene synthase gene GhTPS1 in cotton negatively affects multiple pests while attracting parasitoids. Pest Manag. Sci. 2019. [Google Scholar] [CrossRef]
- Huang, X.Z.; Xiao, Y.T.; Kollner, T.G.; Jing, W.X.; Kou, J.F.; Chen, J.Y.; Liu, D.F.; Gu, S.H.; Wu, J.X.; Zhang, Y.J. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018, 41, 261–274. [Google Scholar] [CrossRef]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Effects of Cucumber mosaic virus infection on vector and non-vector herbivores of squash. Commun. Integr. Biol. 2010, 3, 579–582. [Google Scholar] [CrossRef]
- Carr, J.P.; Tungadi, T.; Donnelly, R.; Bravo-Cazar, A.; Rhee, S.J.; Watt, L.G.; Mutuku, J.M.; Wamonje, F.O.; Murphy, A.M.; Arinaitwe, W.; et al. Modelling and manipulation of aphid-mediated spread of non-persistently transmitted viruses. Virus Res. 2019, 277, 197845. [Google Scholar] [CrossRef]
- Rosen, R.; Kanakala, S.; Kliot, A.; Cathrin Pakkianathan, B.; Farich, B.A.; Santana-Magal, N.; Elimelech, M.; Kontsedalov, S.; Lebedev, G.; Cilia, M.; et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 2015, 15, 1–8. [Google Scholar] [CrossRef]
- Wang, X.W.; Li, P.; Liu, S.S. Whitefly interactions with plants. Curr. Opin. Insect Sci. 2017, 19, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Belliure, B.; Janssen, A.; Sabelis, M.W. Herbivore benefits from vectoring plant virus through reduction of period of vulnerability to predation. Oecologia 2008, 156, 797–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Biochemical and physiological mechanisms underlying effects of Cucumber mosaic virus on host-plant traits that mediate transmission by aphid vectors. Plant Cell Environ. 2014, 37, 1427–1439. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yoshida, Y. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–e10777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, K.; Peng, Y.J.; Yuan, L.B.; Dai, Y.S.; Chen, Q.F.; Yu, L.J.; Bai, M.Y.; Zhang, W.Q.; Xie, L.J.; Xiao, S. Brassinosteroids Antagonize Jasmonates-Activated Plant Defense Responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 2019. [Google Scholar] [CrossRef]
- Hou, X.; Ding, L.; Yu, H. Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 2013, 32, 1067–1074. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, M.; Gangappa, S.N.; Maurya, J.P.; Sethi, V.; Srivastava, A.K.; Singh, A.; Dutta, S.; Ojha, M.; Gupta, N.; Sengupta, M.; et al. Functional interrelation of MYC2 and HY5 plays an important role in Arabidopsis seedling development. Plant J. 2019, 99, 1080–1097. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Pierik, R.; Van Wees, S.C. Different shades of JAZ during plant growth and defense. New Phytol. 2014, 204, 261–264. [Google Scholar] [CrossRef]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Oliveira Ferreira, D.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Chen, F.; Mannas, J.P.; Feldman, T.; Sumner, L.W.; Roossinck, M.J. Virus infection improves drought tolerance. New Phytol. 2008, 180, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; McCann, L.; Naish, M.; Dixon, H.; Murphy, A.M.; Stancombe, M.A.; Bennett, M.H.; Powell, G.; Webb, A.A.; Carr, J.P. A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol. Plant Pathol. 2013, 14, 158–170. [Google Scholar] [CrossRef]
- Carr, J.P.; Donnelly, R.; Tungadi, T.; Murphy, A.M.; Jiang, S.; Bravo-Cazar, A.; Yoon, J.Y.; Cunniffe, N.J.; Glover, B.J.; Gilligan, C.A. Viral Manipulation of Plant Stress Responses and Host Interactions With Insects. Adv Virus Res. 2018, 102, 177–197. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, X.; Yao, X.; Zhang, P. A 7-Amino-Acid Motif of Rep Protein Essential for Virulence Is Critical for Triggering Host Defense Against Sri Lankan Cassava Mosaic Virus. Mol Plant Microbe Interact. 2020, 33, 78–86. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Xu, R.; Qu, J.; Tee, C.; Jiang, X.; Ye, J. DNA-A of a highly pathogenic Indian cassava mosaic virus isolated from Jatropha curcas causes symptoms in Nicotiana benthamiana. Virus Genes. 2014, 48, 402–405. [Google Scholar] [CrossRef]
- Heck, M.; Brault, V. Targeted disruption of aphid transmission: A vision for the management of crop diseases caused by Luteoviridae members. Curr. Opin. Virol. 2018, 33, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.E.; Whitfield, A.E. The Genus Tospovirus: Emerging Bunyaviruses that Threaten Food Security. Annu. Rev. Virol. 2016, 3, 101–124. [Google Scholar] [CrossRef]
- Tungadi, T.; Donnelly, R.; Qing, L. Cucumber mosaic virus 2b proteins inhibit virus-induced aphid resistance in tobacco. Mol. Plant Pathol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Qu, J.; Mao, H.Z.; Ma, Z.G.; Rahman, N.E.; Bai, C.; Chen, W.; Jiang, S.Y.; Ramachandran, S.; Chua, N.H. Engineering geminivirus resistance in Jatropha curcus. Biotechnol. Biofuels 2014, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worrall, E.A.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.P.; Mitter, N. Exogenous Application of RNAi-Inducing Double-Stranded RNA Inhibits Aphid-Mediated Transmission of a Plant Virus. Front Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Bing, X.L.; Li, M.; Ye, G.Y.; Liu, S.S. Infection of tobacco plants by a begomovirus improves nutritional assimilation by a whitefly. Entomol. Exp. Appl. 2012, 144, 191–201. [Google Scholar] [CrossRef]
- Luan, J.B.; Wang, X.W.; Colvin, J.; Liu, S.S. Plant-mediated whitefly–begomovirus interactions: Research progress and future prospects. Bull. Entomol. Res. 2014, 104, 267–276. [Google Scholar] [CrossRef]
- Jiu, M.; Zhou, X.P.; Tong, L.; Xu, J.; Yang, X.; Wan, F.H.; Liu, S.S. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2007, 2, e182. [Google Scholar] [CrossRef] [Green Version]
- Long, E.Y.; Finke, D.L. Predators indirectly reduce the prevalence of an insect-vectored plant pathogen independent of predator diversity. Oecologia 2015, 177, 1067–1074. [Google Scholar] [CrossRef]
- Groen, S.C.; Jiang, S.; Murphy, A.M.; Cunniffe, N.J.; Westwood, J.H.; Davey, M.P.; Bruce, T.J.; Caulfield, J.C.; Furzer, O.J.; Reed, A.; et al. Virus Infection of Plants Alters Pollinator Preference: A Payback for Susceptible Hosts? PLoS Pathog. 2016, 12, e1005790. [Google Scholar] [CrossRef] [Green Version]
| Species | Effectors-Plant Targets | Mechanism | Reference |
|---|---|---|---|
| Virus | |||
| Tomato yellow leaf curl China virus (begomovirus) | βC1-MYC2 | Subvert defense gene activity, compromise terpene synthase | [77] |
| Tomato yellow leaf curl virus (begomovirus) | C2-JAZ1 | Compromise JAZ1 degradation, inhibit downstream gene regulated defense | [78] |
| Tomato spotted wilt orthotospovirus (tospovirus) | NSs-MYC2 | Directly interact with MYCs to disable JA-mediated host defenses against the thrip vector | [79] |
| Cucumber mosaic virus (bromovirus) | 2b-JAZ | Repress the JA-induced degradation of JAZ proteins | [80] |
| Bacterium and Fungal | |||
| Pseudomonas syringae pv. tabaci (Pta) 11528 | HopX1-JAZ | Encode a cysteine protease to promote the degradation of JAZ proteins | [81] |
| Pseudomonas syringae strain A2 | HopZ1a-JAZ | JAZs can be acetylated by HopZ1a through putative acetyltransferase activity. | [82] |
| Pseudomonas syringae | AvrB-JAZ | Induce the COI1–JAZs interactions and the degradation of multiple JAZ proteins | [83] |
| Laccaria bicolor | MiSSP7-JAZ | Interact with and stabilize JAZ proteins against JA-Ile mediated degradation | [84] |
| Pseudomonas syringae | HopBB1-JAZ | Utilize JAZ3 to target TCP14 to the SCFCOI1 degradation complex | [85] |
| Pseudomonas fluorescens WCS417r | ? /-MYC2 | Utilize MYC2 to enhance JA-mediated induced systemic resistance against pathogens and insect herbivores | [86] |
| Pseudomonas syringae | COR-COI1/JAZ | Depredate JAZ, activate the host’s JA signaling pathway to suppress salicylic acid to promote bacterial virulence | [87] |
| Insect | |||
| Helicoverpa armigera | HARP1-JAZ | Directly interact with JAZ to prevent its COI1-mediated degradation | [70] |
| Pieris brassicae | egg extract-MYC | Diminished MYC protein levels in a SA-dependent manner | [57] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Ye, J. Manipulation of Jasmonate Signaling by Plant Viruses and Their Insect Vectors. Viruses 2020, 12, 148. https://0-doi-org.brum.beds.ac.uk/10.3390/v12020148
Wu X, Ye J. Manipulation of Jasmonate Signaling by Plant Viruses and Their Insect Vectors. Viruses. 2020; 12(2):148. https://0-doi-org.brum.beds.ac.uk/10.3390/v12020148
Chicago/Turabian StyleWu, Xiujuan, and Jian Ye. 2020. "Manipulation of Jasmonate Signaling by Plant Viruses and Their Insect Vectors" Viruses 12, no. 2: 148. https://0-doi-org.brum.beds.ac.uk/10.3390/v12020148





