In Silico Discovery of Candidate Drugs against Covid-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Public Datasets
2.2. Correlation, Gene Ontology and Enrichment Analysis
2.3. Protein-Protein and Drug Interaction
3. Results
4. Discussion
4.1. Gene Ontology and Pathway Enrichment Analysis
4.2. High Degree Centrality Gene Study and Potential Drug Therapy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. SARS (Severe Acute Respiratory Syndrome). 2004. Available online: https://www.who.int/ith/diseases/sars/en/ (accessed on 4 April 2020).
- Huang, L.L.; Shen, S.P.; Yu, P.; Wei, Y.Y. Dynamic basic reproduction number based evaluation for current prevention and control of COVID-19 outbreak in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Chen, Y.; Qin, Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.B.; Zhong, C.K.; Zhang, K.X.; Dong, C.; Peng, H.; Xu, T.; Wang, A.L.; Guo, Z.R.; Zhang, Y.H. Epidemic trend of corona virus disease 2019 (COVID-19) in mainland China. Zhonghua Yu Fang Yi Xue Za Zhi 2020, 54, E022. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Coronavirus Disease 2019 (COVID-19) Situation Report –43. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 4 April 2020).
- Xu, J.; Shi, P.Y.; Li, H.; Zhou, J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Fan, H.H.; Wang, L.Q.; Liu, W.L.; An, X.P.; Liu, Z.D.; He, X.Q.; Song, L.H.; Tong, Y.G. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin. Med. J. (Engl). 2020. [Google Scholar] [CrossRef]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Dinnon, K.H.; Yount, B.L., Jr.; McAnarney, E.T.; Gralinski, L.E.; Hale, A.; Graham, R.L.; Scobey, T.; Anthony, S.J.; Wang, L.; et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronavirus Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H. Pulmonary Angiotensin-Converting Enzyme 2 (ACE2) and Inflammatory Lung Disease. Shock 2016, 46, 239–248. [Google Scholar] [CrossRef]
- Burrell, L.M.; Johnston, C.I.; Tikellis, C.; Cooper, M.E. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol Metab. 2004, 15, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.C.; Li, Y.M.; Ma, J.L.; Yi, N.; Yao, Z.Y.; Li, Y.P.; Quan, Y.; Li, X.N.; Xu, C.L.; Qiu, Y.; et al. Single-cell RNA sequencing reveals distinct gene expression patterns in glucose metabolism of human preimplantation embryos. Reprod Fertil Dev. 2019, 31, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Murray, J.L.; Zhao, J.; Sheng, J.; Zhao, Z.; Rubin, D.H. Systems Biology-Based Investigation of Cellular Antiviral Drug Targets Identified by Gene-Trap Insertional Mutagenesis. PLoS Comput. Biol. 2016, 12, e1005074. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Murray, J.L.; Rubin, D.H. Drug Repurposing: New Treatments for Zika Virus Infection? Trends Mol. Med. 2016, 22, 919–921. [Google Scholar] [CrossRef]
- Mustafa, S.; Balkhy, H.; Gabere, M. Peptide-Protein Interaction Studies of Antimicrobial Peptides Targeting Middle East Respiratory Syndrome Coronavirus Spike Protein: An In Silico Approach. Adv Bioinform. 2019, 2019, 6815105. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Cava, C.; Bertoli, G.; Castiglioni, I. Portrait of Tissue-Specific Coexpression Networks of Noncoding RNAs (miRNA and lncRNA) and mRNAs in Normal Tissues. Comput. Math. Methods Med. 2019, 2019, 9029351. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cava, C.; Colaprico, A.; Bertoli, G.; Bontempi, G.; Mauri, G.; Castiglioni, I. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinform. 2016, 17, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravatà, V.; Cava, C.; Minafra, L.; Cammarata, F.P.; Russo, G.; Gilardi, M.C.; Castiglioni, I.; Forte, G.I. Radiation-Induced Gene Expression Changes in High and Low Grade Breast Cancer Cell Types. Int. J. Mol. Sci. 2018, 19, E1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cava, C.; Colaprico, A.; Bertoli, G.; Graudenzi, A.; Silva, T.C.; Olsen, C.; Noushmehr, H.; Bontempi, G.; Mauri, G.; Castiglioni, I. SpidermiR: An R/Bioconductor Package for Integrative Analysis with miRNA Data. Int. J. Mol. Sci. 2017, 18, E274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cava, C.; Bertoli, G.; Castiglioni, I. In silico identification of drug target pathways in breast cancer subtypes using pathway cross-talk inhibition. J. Transl. Med. 2018, 16, 154. [Google Scholar] [CrossRef]
- Günther, S.; Kuhn, M.; Dunkel, M.; Campillos, M.; Senger, C.; Petsalaki, E.; Ahmed, J.; Urdiales, E.G.; Gewiess, A.; Jensen, L.J.; et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008, 36, D919–D922. [Google Scholar] [CrossRef]
- Cotto, K.C.; Wagner, A.H.; Feng, Y.Y.; Kiwala, S.; Coffman, A.C.; Spies, G.; Wollam, A.; Spies, N.C.; Griffith, O.L.; Griffith, M. DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018, 46, D1068–D1073. [Google Scholar] [CrossRef] [Green Version]
- Nikonova, E.V.; Xiong, Y.; Tanis, K.Q.; Dawson, V.L.; Vogel, R.L.; Finney, E.M.; Stone, D.J.; Reynolds, I.J.; Kern, J.T.; Dawson, T.M. Transcriptional responses to loss or gain of function of the leucine-rich repeat kinase 2 (LRRK2) gene uncover biological processes modulated by LRRK2 activity. Hum. Mol. Genet. 2012, 21, 163–174. [Google Scholar] [CrossRef]
- Václavíková, R.; Hughes, D.J.; Souček, P. Microsomal epoxide hydrolase 1 (EPHX1): Gene, structure, function, and role in human disease. Gene. 2015, 571, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ioannidis, I.; McNally, B.; Willette, M.; Peeples, M.E.; Chaussabel, D.; Durbin, J.E.; Ramilo, O.; Mejias, A.; Flaño, E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J. Virol. 2012, 86, 5422–5436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, J.; Drake, M.J.; Bruce, E.A.; Riblett, A.M.; Didigu, C.A.; Wilen, C.B.; Malani, N.; Male, F.; Lee, F.H.; Bushman, F.D.; et al. The major cellular sterol regulatory pathway is required for Andes virus infection. PLoS Pathog. 2014, 10, e1003911. [Google Scholar] [CrossRef] [PubMed]
- Wudiri, G.A.; Pritchard, S.M.; Li, H.; Liu, J.; Aguilar, H.C.; Gilk, S.D.; Nicola, A.V. Molecular requirement for sterols in herpes simplex virus entry and infectivity. J. Virol. 2014, 88, 13918–13922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Rajendran, R.; Zhao, Y.; Tan, B.; Wu, G.; Bazer, F.W.; Zhu, G.; Peng, Y.; Huang, X.; Deng, J.; et al. Amino Acids As Mediators of Metabolic Cross Talk between Host and Pathogen. Front. Immunol. 2018, 9, 319. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W. The unfolded protein response in virus infections. Front. Microbiol. 2014, 5, 518. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.; Grose, C. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J. Virol. 1988, 62, 2701–2711. [Google Scholar] [CrossRef] [Green Version]
- Duygu, F.; Tekin Koruk, S.; Aksoy, N. Serum paraoxonase and arylesterase activities in various forms of hepatitis B virus infection. J. Clin. Lab. Anal. 2011, 25, 311–316. [Google Scholar] [CrossRef]
- Keller, B.T.; Borchardt, R.T. Adenosine dialdehyde: A potent inhibitor of vaccinia virus multiplication in mouse L929 cells. Mol. Pharmacol. 1987, 31, 485–492. [Google Scholar]
- Abraham, R.; Hauer, D.; McPherson, R.L.; Utt, A.; Kirby, I.T.; Cohen, M.S.; Merits, A.; Leung, A.K.L.; Griffin, D.E. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E10457–E10466. [Google Scholar] [CrossRef] [Green Version]
- Vysochan, A.; Sengupta, A.; Weljie, A.M.; Alwine, J.C.; Yu, Y. ACSS2-mediated acetyl-CoA synthesis from acetate is necessary for human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 2017, 114, E1528–E1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagavan, N.V. Protein and Amino Acid Metabolism. In Medichale Biochemistry, Fourth Edition; Academic Press: San Diego, CA, USA, 2002; pp. 331–363. [Google Scholar]
- Meher, G.; Bhattacharjya, S.; Chakraborty, H. Membrane Cholesterol Modulates Oligomeric Status and Peptide-Membrane Interaction of Severe Acute Respiratory Syndrome Coronavirus Fusion Peptide. J. Phys. Chem. B. 2019, 123, 10654–10662. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, B.; Cho, S.Y.; Oh, K.S.; Kim, S.H.; Kim, Y.O.; Jeong, E.H.; Nguyen, T.T.; Kim, S.H.; Kim, I.S.; Kwon, J.; et al. Effectiveness of Periodic Treatment of Quercetin against Influenza A Virus H1N1 through Modulation of Protein Expression. J. Agric. Food Chem. 2016, 64, 4416–4425. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, R.; Kuroda, K.; Yoshida, R.; Tsuda, Y.; Fujikura, D.; Miyamoto, H.; Kajihara, M.; Kida, H.; Takada, A. Heat shock protein 70 modulates influenza A virus polymerase activity. J. Biol. Chem. 2014, 289, 7599–7614. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhuang, L.; Szatmary, P.; Wen, L.; Sun, H.; Lu, Y.; Xu, Q.; Chen, X. Upregulation of heat shock proteins (HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is associated with poor outcomes from HBV-related early-stage hepatocellular carcinoma. Int. J. Med. Sci. 2015, 12, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Osseman, Q.; Gallucci, L.; Au, S.; Cazenave, C.; Berdance, E.; Blondot, M.L.; Cassany, A.; Bégu, D.; Ragues, J.; Aknin, C.; et al. The chaperone dynein LL1 mediates cytoplasmic transport of empty and mature hepatitis B virus capsids. J. Hepatol. 2018, 68, 441–448. [Google Scholar] [CrossRef]
- Esfandiarei, M.; Suarez, A.; Amaral, A.; Si, X.; Rahmani, M.; Dedhar, S.; McManus, B.M. Novel role for integrin-linked kinase in modulation of coxsackievirus B3 replication and virus-induced cardiomyocyte injury. Circ. Res. 2006, 99, 354–361. [Google Scholar] [CrossRef] [Green Version]
- AlQarni, S.; Al-Sheikh, Y.; Campbell, D.; Drotar, M.; Hannigan, A.; Boyle, S.; Herzyk, P.; Kossenkov, A.; Armfield, K.; Jamieson, L.; et al. Lymphomas driven by Epstein-Barr virus nuclear antigen-1 (EBNA1) are dependant upon Mdm2. Oncogene 2018, 37, 3998–4012. [Google Scholar] [CrossRef] [Green Version]
- Park, D.E.; Cheng, J.; Berrios, C.; Montero, J.; Cortés-Cros, M.; Ferretti, S.; Arora, R.; Tillgren, M.L.; Gokhale, P.C.; DeCaprio, J.A. Dual inhibition of MDM2 and MDM4 in virus-positive Merkel cell carcinoma enhances the p53 response. Proc. Natl Acad Sci USA. 2019, 116, 1027–1032. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Kariyawasam, H.H.; Scadding, G.K. Seasonal allergic rhinitis: Fluticasone propionate and fluticasone furoate therapy evaluated. J. Asthma Allergy. 2010, 3, 19–28. [Google Scholar] [PubMed]
- Staresinic, A.G.; Sorkness, C.A. Fluticasone propionate: A potent inhaled corticosteroid for the treatment of asthma. Expert. Opin. Pharmacother. 2000, 1, 1227–1244. [Google Scholar] [CrossRef] [PubMed]
- van Arman, G.G.; Campbell, W.C. Anti-inflammatory activity of thiabendazole and its relation to parasitic disease. Tex. Rep. Biol. Med. 1975, 33, 303–311. [Google Scholar] [PubMed]
- Wiehe, A.; O’Brien, J.M.; Senge, M.O. Trends and targets in antiviral phototherapy. Photochem. Photobiol. Sci. 2019, 18, 2565–2612. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.M.; Balfour, J.A. Didanosine. An update on its antiviral activity, pharmacokinetic properties and therapeutic efficacy in the management of HIV disease. Drugs 1996, 52, 928–962. [Google Scholar] [CrossRef]
- Baliji, S.; Lacatus, G.; Sunter, G. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 2010, 402, 238–247. [Google Scholar] [CrossRef] [Green Version]
- Dasinger, J.H.; Intapad, S.; Rudsenske, B.R.; Davis, G.K.; Newsome, A.D.; Alexander, B.T. Chronic Blockade of the Androgen Receptor Abolishes Age-Dependent Increases in Blood Pressure in Female Growth-Restricted Rats. Hypertension 2016, 67, 1281–1290. [Google Scholar] [CrossRef] [Green Version]
- Mishra, J.S.; Hankins, G.D.; Kumar, S. Testosterone downregulates angiotensin II type-2 receptor via androgen receptor-mediated ERK1/2 MAP kinase pathway in rat aorta. J. Renin. Angiotensin Aldosterone Syst. 2016, 17. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jiang., Q.; Xia, X.; Liu, K.; Yu, Z.; Tao, W.; Gong, W.; Han, J.J. Individual Variation of the SARS-CoV2 Receptor ACE2 Gene Expression and Regulation. Preprints 2020, 2020030191. [Google Scholar]

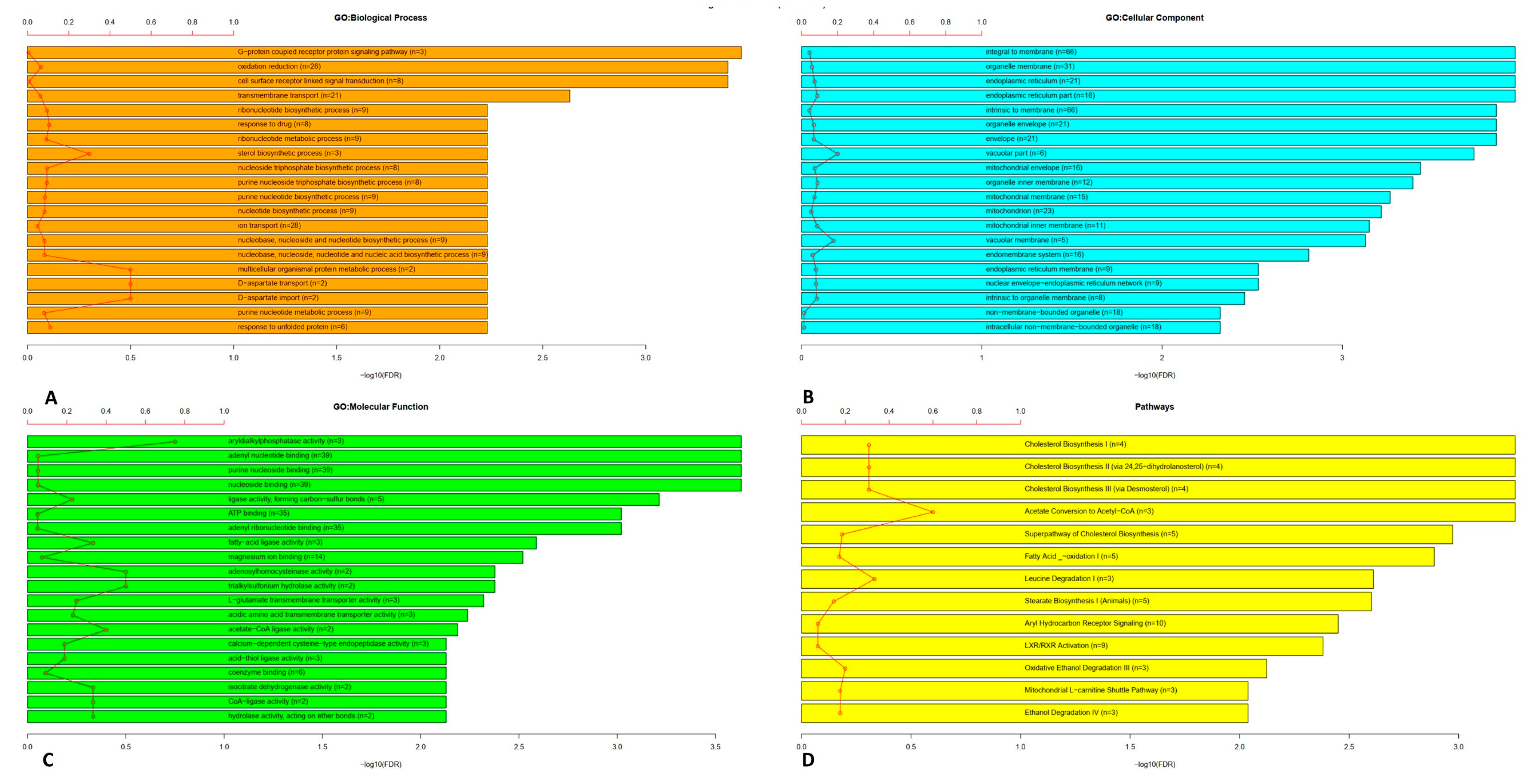
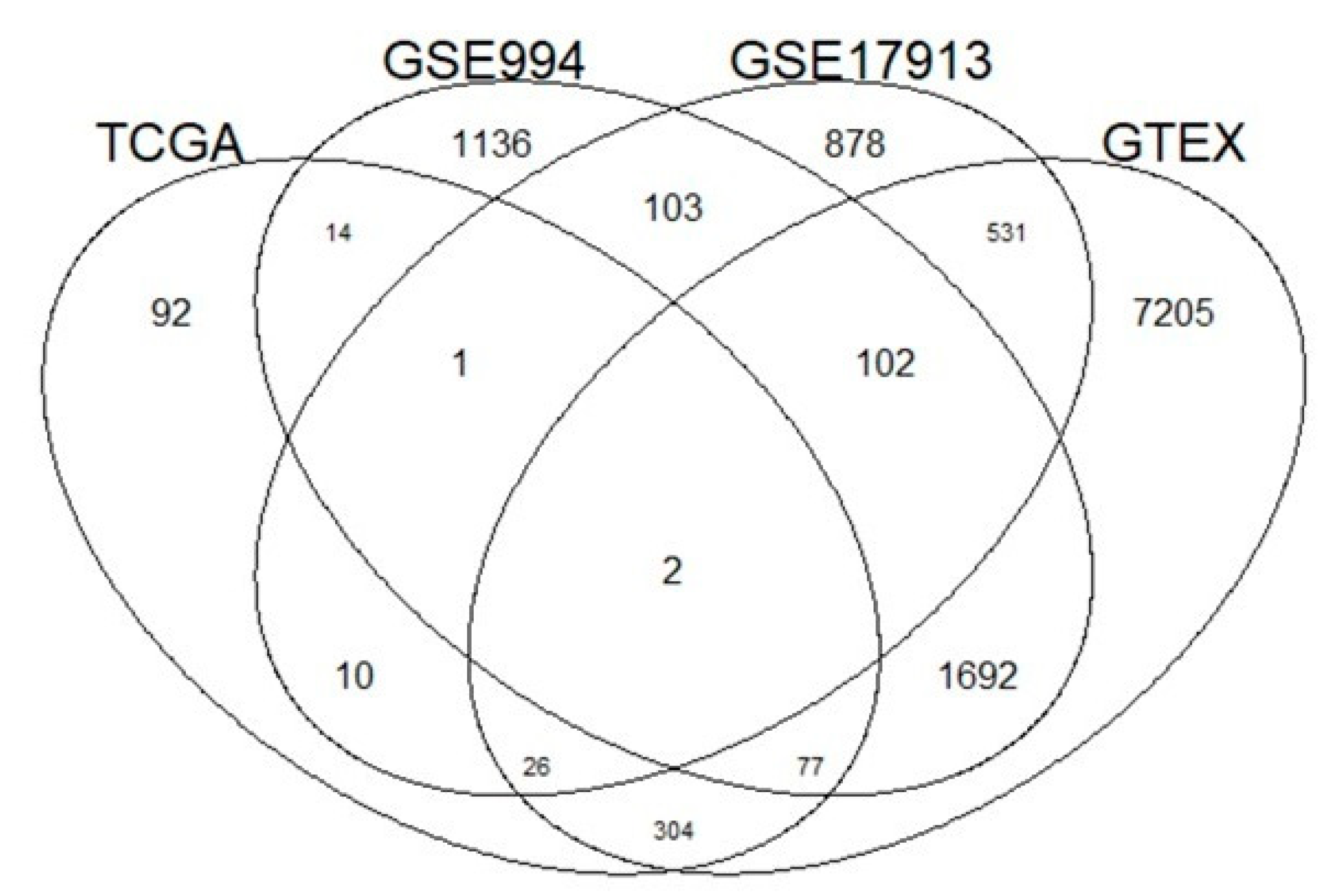
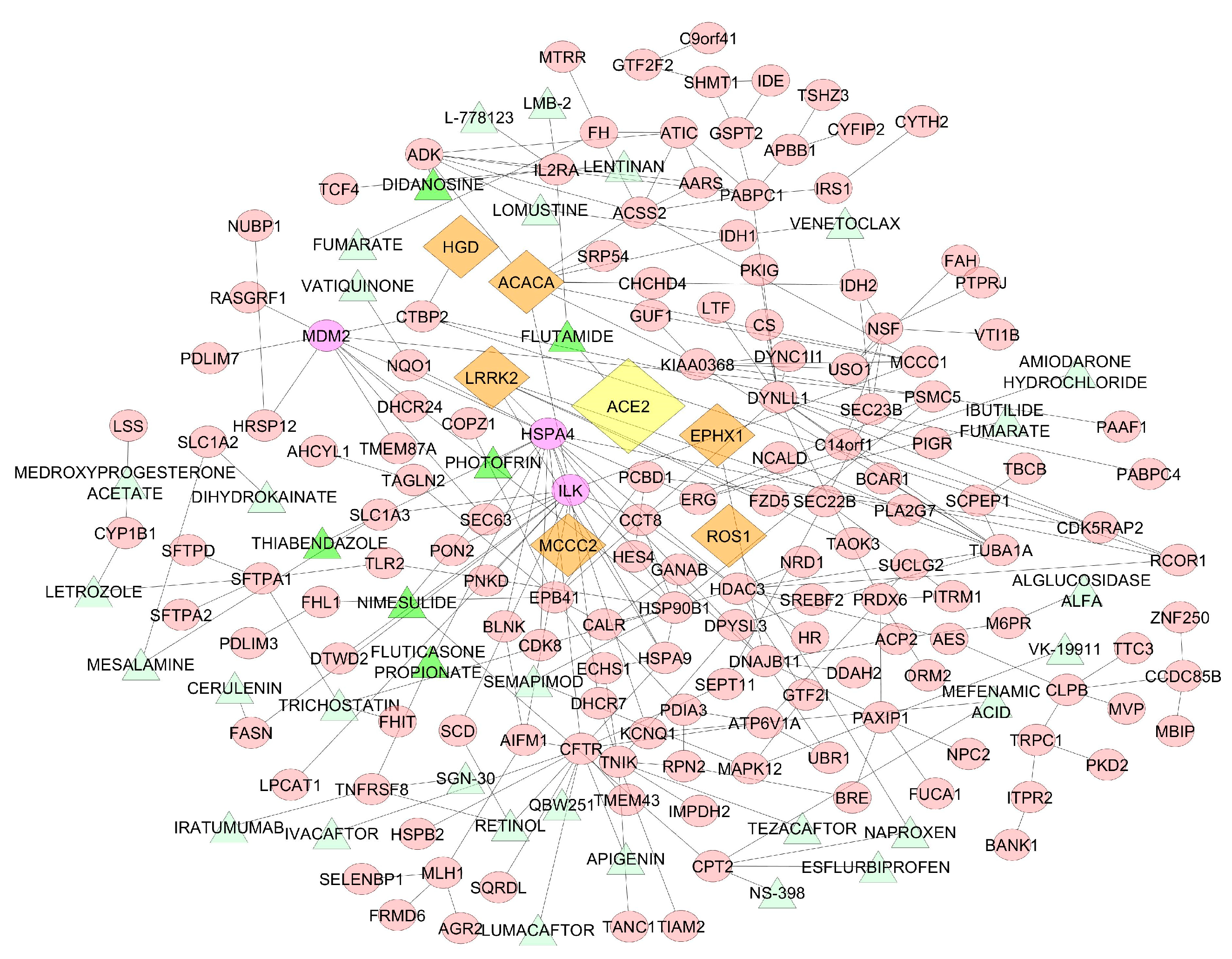
| Name | Function | Gene Ontology | Drug | p-value | Correlation |
|---|---|---|---|---|---|
| Leucine-Rich Repeat Kinase 2 (LRRK2) | It is involved in multiple activities such as neuronal plasticity, autophagy, and vesicle trafficking | MAPK cascade | Tamoxifen | 2 × 10−8 | 0.66 |
| Acyl-CoA Synth. Long Chain Family Memb 5 (ACSL5) | It participates in lipid biosynthesis and fatty acid degradation | long-chain fatty acid metabolic process | 3 × 10−8 | 0.65 | |
| Cysteine Rich Protein 2 (CRIP2) | It is involved in the differentiation of smooth muscle tissue | protein binding | 5 × 10−8 | −0.64 | |
| Hydroxysteroid 17-Beta Dehydrogen. 4 (HSD17B4) | It plays a role in the peroxisomal beta-oxidation pathway for fatty acids | very long-chain fatty acid metabolic process | 1 × 10−7 | 0.63 | |
| Epoxide Hydrolase 1 (EPHX1) | It participates in the metabolism of lipids | epoxide hydrolase activity | Carbamazepine, Clofibrate, Phenobarbital, AR9281 | 4 × 10−7 | 0.60 |
| Methylcrotonoyl-CoA Carboxylase 2 (MCCC2) | It is involved in the leucine and isovaleric acid catabolism | protein binding | 7 × 10−7 | 0.60 | |
| Glutathione S-Transferase Alpha 4 (GSTA4) | It plays a role in cellular defense against oxidative stress | glutathione transferase activity | 9 × 10−7 | 0.59 | |
| Acetyl-CoA Carboxylase Alpha (ACACA) | It participates in fatty acid synthesis | tissue homeostasis | metformin | 4 × 10−6 | 0.56 |
| Homogentisate 1,2-Dioxygenase (HGD) | It plays a role in the catabolism of the amino acids | protein binding | 5 × 10−6 | 0.56 | |
| ROS Proto-Oncogene 1, Rec. Tyros. Kinase (ROS1) | It contributes in epithelial cell differentiation | regulation of cell growth | Crizotinib Brigatinib Entrectinib Cabozantinib Ceritinib Lorlatinib Foretinib Naproxen Asp-3026 Tae-684 Imatinib | 6 × 10−6 | 0.55 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cava, C.; Bertoli, G.; Castiglioni, I. In Silico Discovery of Candidate Drugs against Covid-19. Viruses 2020, 12, 404. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040404
Cava C, Bertoli G, Castiglioni I. In Silico Discovery of Candidate Drugs against Covid-19. Viruses. 2020; 12(4):404. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040404
Chicago/Turabian StyleCava, Claudia, Gloria Bertoli, and Isabella Castiglioni. 2020. "In Silico Discovery of Candidate Drugs against Covid-19" Viruses 12, no. 4: 404. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040404





