Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Type of Study
2.2. Ethical Considerations
2.3. Location
2.4. Sampling, Collection, and Storage
2.5. RNA Extraction and Complementary DNA Synthesis
2.6. Polymersase Chain Reaction (PCR) and Sequencing
2.7. Sequencing and Phylogenetic Analysis
2.8. Statistical Analysis
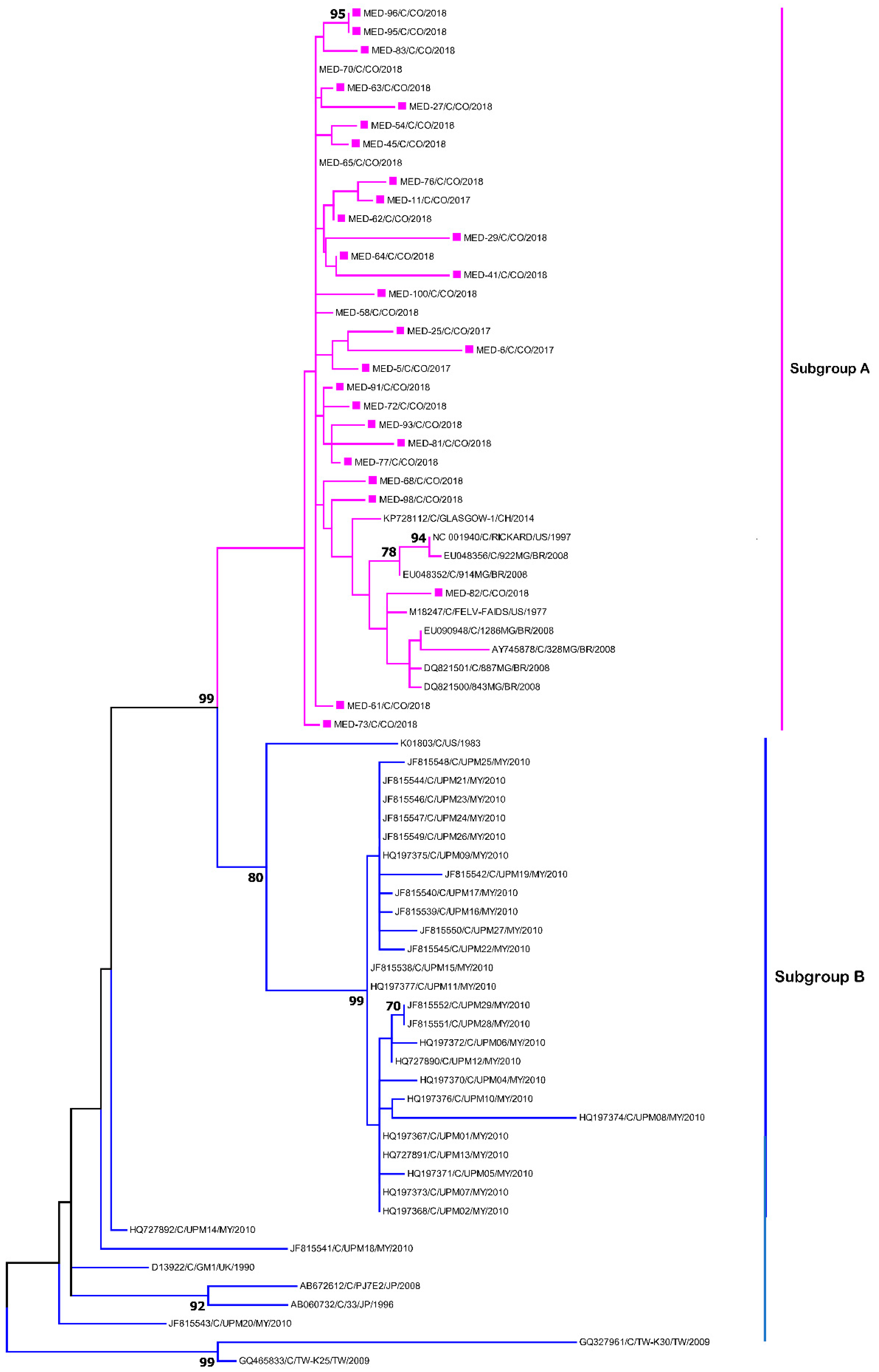
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunham, S.P.; Graham, E. Retroviral infections of small animals. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 879–901. [Google Scholar] [CrossRef] [PubMed]
- Cattori, V.; Tandon, R.; Riond, B.; Pepin, A.C.; Lutz, H.; Hofmann-Lehmann, R. The kinetics of feline leukaemia virus shedding in experimentally infected cats are associated with infection outcome. Vet. Microbiol. 2009, 133, 292–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benveniste, R.E.; Sherr, C.J.; Todaro, G.J. Evolution of type C viral genes: Origin of feline leukemia virus. Science 1975, 190, 886–888. [Google Scholar] [CrossRef] [PubMed]
- Chiu, E.S.; Hoover, E.A.; VandeWoude, S. A Retrospective Examination of Feline Leukemia Subgroup Characterization: Viral Interference Assays to Deep Sequencing. Viruses 2018, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Willett, B.J.; Hosie, M.J. Feline leukaemia virus: Half a century since its discovery. Vet. J. 2013, 195, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Sheets, R.L.; Pandey, R.; Klement, V.; Grant, C.K.; Roy-burman, P. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology 1992, 190, 849–855. [Google Scholar] [CrossRef]
- Stewart, H.; Adema, K.W.; McMonagle, E.L.; Hosie, M.J.; Willett, B.J. Identification of novel subgroup A variants with enhanced receptor binding and replicative capacity in primary isolates of anaemogenic strains of feline leukaemia virus. Retrovirology 2012, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Anai, Y.; Ochi, H.; Watanabe, S.; Nakagawa, S.; Kawamura, M.; Gojobori, T.; Nishigaki, K. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J. Virol. 2012, 86, 8634–8644. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.M.; Lauring, A.S.; Burns, C.C.; Overbaugh, J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 2000, 287, 1828–1830. [Google Scholar] [CrossRef]
- Miyake, A.; Watanabe, S.; Hiratsuka, T.; Ito, J.; Ngo, M.H.; Makundi, I.; Kawasaki, J.; Endo, Y.; Tsujimoto, H.; Nishigaki, K. Novel Feline Leukemia Virus Interference Group Based on the env Gene. J. Virol. 2016, 90, 4832–4837. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, K. Clinical aspects of feline retroviruses: A review. Viruses 2012, 4, 2684–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz, L. Haemopoyetic neoplasm in 10 cats positive to feline leukaemia virus. Arch. Med. Vet. 2005, 37, 71–76. [Google Scholar] [CrossRef]
- Belgard, S.; Truyen, U.; Thibault, J.C.; Sauter-Louis, C.; Hartmann, K. Relevance of feline calicivirus, feline immunodeficiency virus, feline leukemia virus, feline herpesvirus and Bartonella henselae in cats with chronic gingivostomatitis. Berl. Munch Tierarztl. Wochenschr. 2010, 123, 369–376. [Google Scholar] [PubMed]
- Quimby, J.M.; Elston, T.; Hawley, J.; Brewer, M.; Miller, A.; Lappin, M.R. Evaluation of the association of Bartonella species, feline herpesvirus 1, feline calicivirus, feline leukemia virus and feline immunodeficiency virus with chronic feline gingivostomatitis. J. Feline Med. Surg. 2008, 10, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.A.; Chiu, E.S.; Kraberger, S.J.; Roelke-Parker, M.; Lowery, I.; Erbeck, K.; Troyer, R.; Carver, S.; VandeWoude, S. Feline Leukemia Virus (FeLV) Disease Outcomes in a Domestic Cat Breeding Colony: Relationship to Endogenous FeLV and Other Chronic Viral Infections. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.K.; Scott, H.M.; Lachtara, J.L.; Crawford, P.C. Seroprevalence of feline leukemia virus and feline immunodeficiency virus infection among cats in North America and risk factors for seropositivity. J. Am. Vet. Med. Assoc. 2006, 228, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Benavides, H. Presente y futuro de la leucemia viral felina y del virus de inmunodeficiencia adquirida felina en santa Fe de Bogotá. Mem. Semin. Med. Felina Bogotá 1996, 1999, 114–128. [Google Scholar]
- Quintero Garay, J.G. Estudios preliminares de la Leucemia Viral Felina en Santafe de Bogota; Universidad de la Salle: Bogotá, Colombia, 1994. [Google Scholar]
- Tique, V.; Sánchez, A.; Álvarez, A.; Ríos, R.; Mattar, S. Seroprevalencia del virus de Leucemia e Inmunodeficiencia Felina en gatos de Montería. Córdoba. Med. Vet. Zoot 2009, 56, 85–94. [Google Scholar]
- Calle-Restrepo, J.; Fernandez-Gonzalez, L.; Morales-Zapata, L.; Ruiz-Sáenz, J. Feline leukemia virus: A current pathogen requiring attention in Colombia. Veterinaria Y Zootecnía 2013, 7, 117–138. [Google Scholar]
- Miyazawa, T.; Jarrett, O. Feline leukaemia virus proviral DNA detected by polymerase chain reaction in antigenaemic but non-viraemic (‘discordant’) cats. Arch. Virol. 1997, 142, 323–332. [Google Scholar] [CrossRef]
- Geret, C.P.; Cattori, V.; Meli, M.L.; Riond, B.; Martinez, F.; Lopez, G.; Vargas, A.; Simon, M.A.; Lopez-Bao, J.V.; Hofmann-Lehmann, R.; et al. Feline leukemia virus outbreak in the critically endangered Iberian lynx (Lynx pardinus): High-throughput sequencing of envelope variable region A and experimental transmission. Arch. Virol. 2011, 156, 839–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burling, A.N.; Levy, J.K.; Scott, H.M.; Crandall, M.M.; Tucker, S.J.; Wood, E.G.; Foster, J.D. Seroprevalences of feline leukemia virus and feline immunodeficiency virus infection in cats in the United States and Canada and risk factors for seropositivity. J. Am. Vet. Med. Assoc. 2017, 251, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Collazos Paz, M.A. Coinfección y Hallazgos Epidemiológicos de los Virus de Inmunodeficiencia Felina (VIF) y Leucemia Felina (VILEF) en Gatos Clínicamente Enfermos; Facultad de Ciencias, Universidad Javeriana: Bogotá, Colombia, 2016. [Google Scholar]
- Sivagurunathan, A.; Atwa, A.M.; Lobetti, R. Prevalence of feline immunodeficiency virus and feline leukaemia virus infection in Malaysia: A retrospective study. JFMS Open Rep. 2018, 4, 2055116917752587. [Google Scholar] [CrossRef] [PubMed]
- Gates, M.C.; Vigeant, S.; Dale, A. Prevalence and risk factors for cats testing positive for feline immunodeficiency virus and feline leukaemia virus infection in cats entering an animal shelter in New Zealand. N. Z. Vet. J. 2017, 65, 285–291. [Google Scholar] [CrossRef]
- Luckman, C.; Gates, M.C. Epidemiology and clinical outcomes of feline immunodeficiency virus and feline leukaemia virus in client-owned cats in New Zealand. JFMS Open Rep. 2017, 3, 2055116917729311. [Google Scholar] [CrossRef]
- Studer, N.; Lutz, H.; Saegerman, C.; Gonczi, E.; Meli, M.L.; Boo, G.; Hartmann, K.; Hosie, M.J.; Moestl, K.; Tasker, S.; et al. Pan-European Study on the Prevalence of the Feline Leukaemia Virus Infection—Reported by the European Advisory Board on Cat Diseases (ABCD Europe). Viruses 2019, 11, 993. [Google Scholar] [CrossRef] [Green Version]
- Coelho, F.M.; Bomfim, M.R.; de Andrade Caxito, F.; Ribeiro, N.A.; Luppi, M.M.; Costa, E.A.; Oliveira, M.E.; Da Fonseca, F.G.; Resende, M. Naturally occurring feline leukemia virus subgroup A and B infections in urban domestic cats. J. Gen. Virol. 2008, 89 Pt 11, 2799–2805. [Google Scholar] [CrossRef]
- Poffo, D.; Almeida, A.B.; Nakazato, L.; Dutra, V.; Correa, S.H.; Mendonça, A.J.; Sousa, V.R. Feline immunodeficiency virus (FIV), feline leukaemia virus (FeLV) and Leishmania sp. in domestic cats in the Midwest of Brazil. Pesquisa Veterinária Brasileira 2017, 37, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Lacerda, L.C.; Silva, A.N.; Freitas, J.S.; Cruz, R.D.S.; Said, R.A.; Munhoz, A.D. Feline immunodeficiency virus and feline leukemia virus: Frequency and associated factors in cats in northeastern Brazil. Genet. Mol. Res. 2017, 16, 1–8. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Hassan, L.; Zakaria, Z. Molecular detection, phylogenetic analysis, and identification of transcription motifs in feline leukemia virus from naturally infected cats in malaysia. Vet. Med. Int. 2014, 2014, 760961. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.K.; Crawford, P.C.; Tucker, S.J. Performance of 4 Point-of-Care Screening Tests for Feline Leukemia Virus and Feline Immunodeficiency Virus. J. Vet. Intern. Med. 2017, 31, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Mullins, J.I.; Hoover, E.A.; Overbaugh, J.; Quackenbush, S.L.; Donahue, P.R.; Poss, M.L. FeLV-FAIDS-induced immunodeficiency syndrome in cats. Vet. Immunol. Immunopathol. 1989, 21, 25–37. [Google Scholar] [CrossRef]
- Hoover, E.A.; Ebner, J.P.; Zeidner, N.S.; Mullins, J.I. Early therapy of feline leukemia virus infection (FeLV-FAIDS) with 9-(2-phosphonylmethoxyethyl) adenine (PMEA). Antiviral. Res. 1991, 16, 77–92. [Google Scholar] [CrossRef]
- Overbaugh, J.; Hoover, E.A.; Mullins, J.I.; Burns, D.P.; Rudensey, L.; Quackenbush, S.L.; Stallard, V.; Donahue, P.R. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology 1992, 188, 558–569. [Google Scholar] [CrossRef]
- Rohn, J.L.; Moser, M.S.; Gwynn, S.R.; Baldwin, D.N.; Overbaugh, J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 1998, 72, 2686–2696. [Google Scholar] [CrossRef] [Green Version]
- Phipps, A.J.; Hayes, K.A.; Al-dubaib, M.; Roy-Burman, P.; Mathes, L.E. Inhibition of feline leukemia virus subgroup A infection by coinoculation with subgroup B. Virology 2000, 277, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Neil, J.C.; Fulton, R.; Rigby, M.; Stewart, M. Feline leukaemia virus: Generation of pathogenic and oncogenic variants. Curr. Top Microbiol. Immunol. 1991, 171, 67–93. [Google Scholar]

| Male | Female | AGE Range | Ag FeLV Positive |
|---|---|---|---|
| 23 | 22 | <1 years | 22 |
| 12 | 19 | 1–3 years | 22 |
| 11 | 13 | >3 years | 16 |
| Clinic Sx | Patients /% |
|---|---|
| A/P Healthy | 54 |
| Rough Hair | 11 |
| Anaemia | 3 |
| Underweight | 8 |
| Conjunctivitis | 2 |
| Gingivitis | 8 |
| Diarrhea | 7 |
| Mycosis | 1 |
| Neuropathy | 2 |
| loss of Appetite | 3 |
| lymphoma | 1 |
| Total | 100 |
| Municipality | Samples | Ag FeLV Positive |
|---|---|---|
| Medellin | 64 | 33 |
| Caldas | 14 | 9 |
| Bello | 8 | 6 |
| Envigado | 6 | 6 |
| Itagui | 3 | 2 |
| Santa Helena | 3 | 3 |
| Sabaneta | 2 | 1 |
| 100 | 60 |
| Subgroup | Strain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 6 | 8 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1.MED-68/C/CO/2018 | |||||||||||||||||||||||
| 2. MED-62/C/CO/2018 | 0.006 | |||||||||||||||||||||||
| 3, MED-73/C/CO/2018 | 0.006 | 0.006 | ||||||||||||||||||||||
| 4.MED-61/C/CO/2018 | 0.008 | 0.006 | 0.008 | |||||||||||||||||||||
| 5. MED-98/C/CO/2018 | 0.01 | 0.008 | 0.012 | 0.01 | ||||||||||||||||||||
| 6. MED-96/C/CO/2018 | 0.01 | 0.008 | 0.01 | 0.01 | 0.015 | |||||||||||||||||||
| 7. MED-25/C/CO/2018 | 0.01 | 0.008 | 0.01 | 0.01 | 0.015 | 0.012 | ||||||||||||||||||
| 8.MED-93/C/CO/2018 | 0.012 | 0.01 | 0.012 | 0.012 | 0.017 | 0.014 | 0.014 | |||||||||||||||||
| 9. MED-29/C/CO/2018 | 0.014 | 0.012 | 0.014 | 0.014 | 0.019 | 0.015 | 0.015 | 0.017 | ||||||||||||||||
| 10.MED-82/C/CO/2018 | 0.015 | 0.014 | 0.015 | 0.015 | 0.017 | 0.017 | 0.017 | 0.019 | 0.021 | |||||||||||||||
| 11. EU048352/C/914MG/BR/2008 | 0.014 | 0.015 | 0.015 | 0.017 | 0.015 | 0.019 | 0.019 | 0.021 | 0.023 | 0.017 | ||||||||||||||
| 12.M18247/C/FELV-FAIDS/US/1977 | 0.014 | 0.012 | 0.014 | 0.014 | 0.015 | 0.015 | 0.015 | 0.017 | 0.019 | 0.01 | 0.012 | |||||||||||||
| 13. NC_001940/C/RICKARD/US/1997 | 0.015 | 0.017 | 0.017 | 0.019 | 0.017 | 0.021 | 0.021 | 0.019 | 0.025 | 0.023 | 0.006 | 0.017 | ||||||||||||
| 14. DQ821501/C/887MG/BR/2008 | 0.017 | 0.015 | 0.017 | 0.017 | 0.019 | 0.019 | 0.019 | 0.017 | 0.023 | 0.014 | 0.015 | 0.008 | 0.017 | |||||||||||
| B | 15. HQ197377/C/UPM11/MY/2010 | 0.023 | 0.025 | 0.025 | 0.027 | 0.029 | 0.029 | 0.029 | 0.031 | 0.033 | 0.031 | 0.029 | 0.033 | 0.031 | 0.037 | |||||||||
| 16. K01803/C/US/1983 | 0.025 | 0.023 | 0.025 | 0.025 | 0.031 | 0.027 | 0.027 | 0.029 | 0.031 | 0.029 | 0.035 | 0.031 | 0.037 | 0.035 | 0.027 | |||||||||
| 17. HQ197375/C/UPM09/MY/2010 | 0.025 | 0.027 | 0.027 | 0.029 | 0.031 | 0.031 | 0.027 | 0.033 | 0.035 | 0.033 | 0.031 | 0.035 | 0.033 | 0.039 | 0.002 | 0.029 | ||||||||
| 18. HQ727892/C/UPM14/MY/2010 | 0.029 | 0.027 | 0.025 | 0.025 | 0.035 | 0.031 | 0.027 | 0.033 | 0.033 | 0.037 | 0.039 | 0.035 | 0.041 | 0.039 | 0.041 | 0.039 | 0.042 | |||||||
| 19 JF815550/C/UPM27/MY/2010 | 0.031 | 0.033 | 0.033 | 0.035 | 0.037 | 0.037 | 0.033 | 0.039 | 0.041 | 0.039 | 0.033 | 0.041 | 0.035 | 0.044 | 0.008 | 0.035 | 0.006 | 0.014 | ||||||
| 20. D13922/C/GM1/UK/1990 | 0.041 | 0.042 | 0.039 | 0.041 | 0.042 | 0.046 | 0.042 | 0.048 | 0.048 | 0.044 | 0.042 | 0.042 | 0.048 | 0.046 | 0.052 | 0.054 | 0.054 | 0.058 | 0.06 | |||||
| 21. AB060732/C/33/JP/1996 | 0.051 | 0.048 | 0.046 | 0.046 | 0.052 | 0.052 | 0.052 | 0.052 | 0.052 | 0.058 | 0.056 | 0.056 | 0.058 | 0.06 | 0.064 | 0.06 | 0.066 | 0.069 | 0.068 | 0.041 | ||||
| 22. GQ465833/C/TW-K25/TW/2009 | 0.06 | 0.062 | 0.058 | 0.06 | 0.066 | 0.064 | 0.064 | 0.064 | 0.068 | 0.071 | 0.066 | 0.069 | 0.064 | 0.069 | 0.062 | 0.069 | 0.062 | 0.068 | 0.068 | 0.042 | 0.062 | |||
| 23. GQ327961/C/TW-K30/TW/2009 | 0.1 | 0.098 | 0.098 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.01 | 0.1 | 100 | 0.1 | 0.1 | 0.085 | 0,1 | 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, C.; Valencia, A.C.; Duque-Valencia, J.; Ruiz-Saenz, J. Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia. Viruses 2020, 12, 464. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040464
Ortega C, Valencia AC, Duque-Valencia J, Ruiz-Saenz J. Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia. Viruses. 2020; 12(4):464. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040464
Chicago/Turabian StyleOrtega, Carolina, Alida C. Valencia, July Duque-Valencia, and Julián Ruiz-Saenz. 2020. "Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia" Viruses 12, no. 4: 464. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040464






