Infectious Norovirus Is Chronically Shed by Immunocompromised Pediatric Hosts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Molecular Detection of Norovirus
2.3. In Vitro Human Norovirus Infection
2.4. Statistical Analyses
3. Results
3.1. Cohort Description
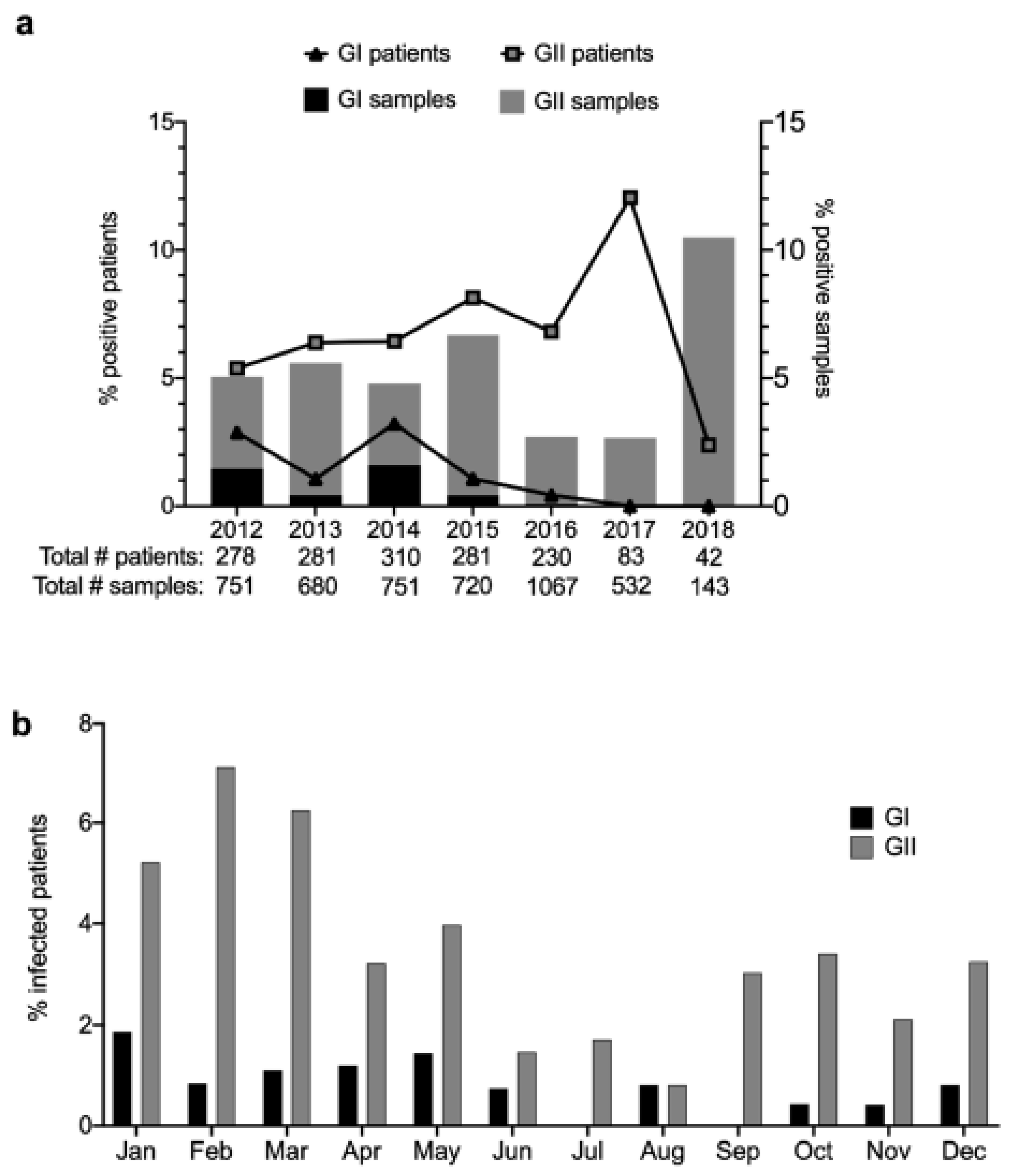
3.2. Prevalence and Temporal Trends of Norovirus Infections
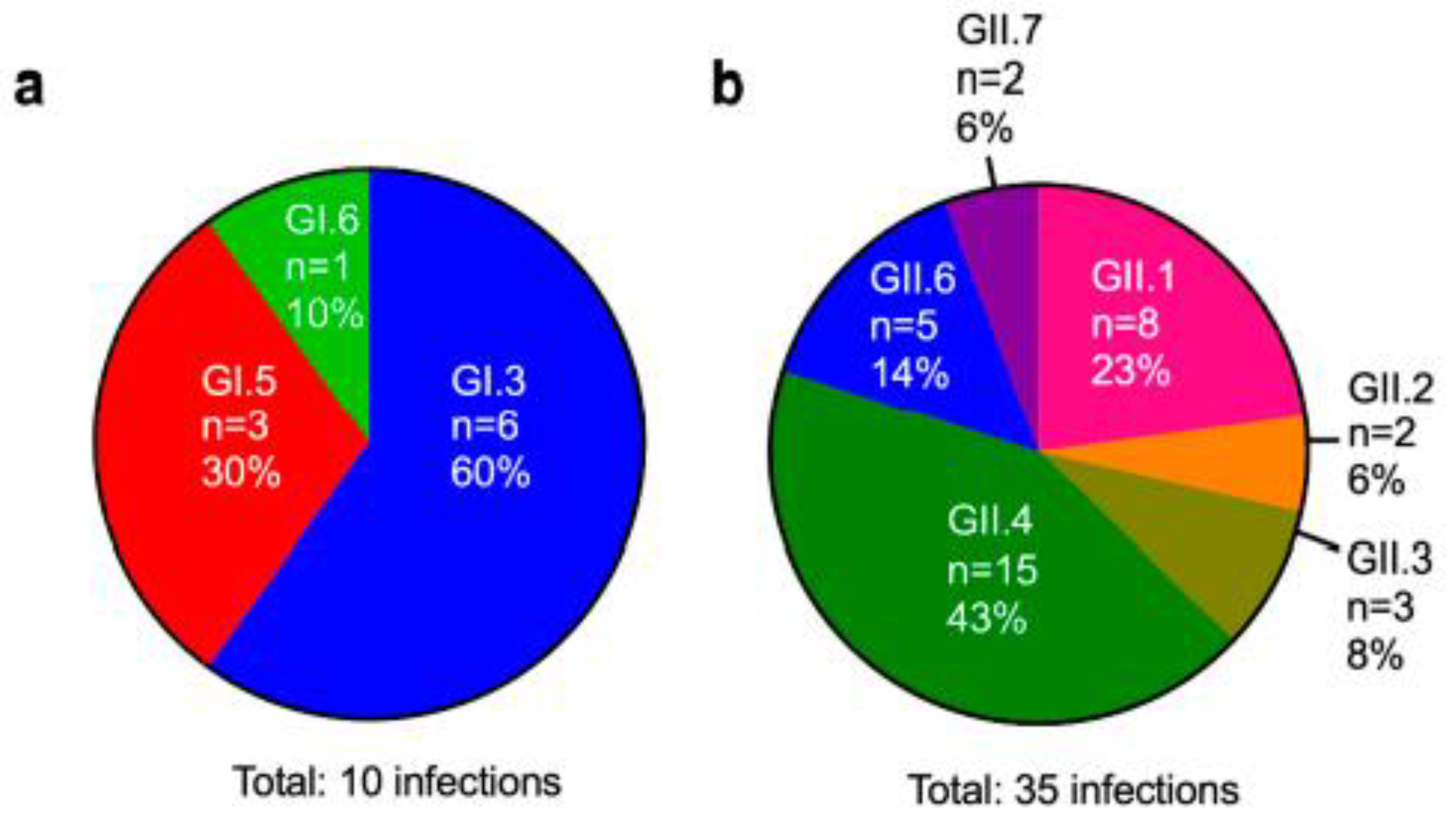
3.3. Genotyping of Patient Isolates
3.4. Persistent Virus Shedding in a Subset of Patients
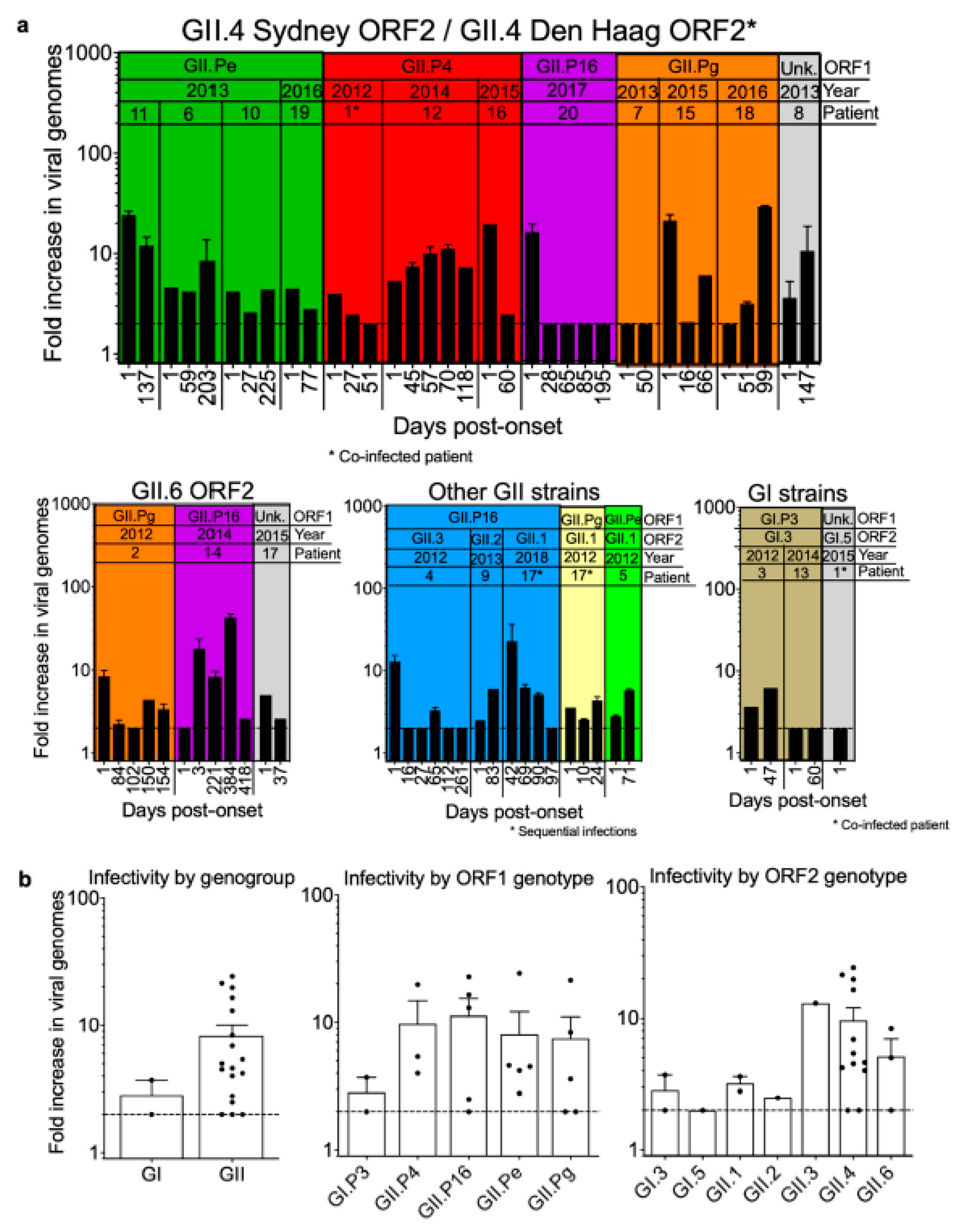
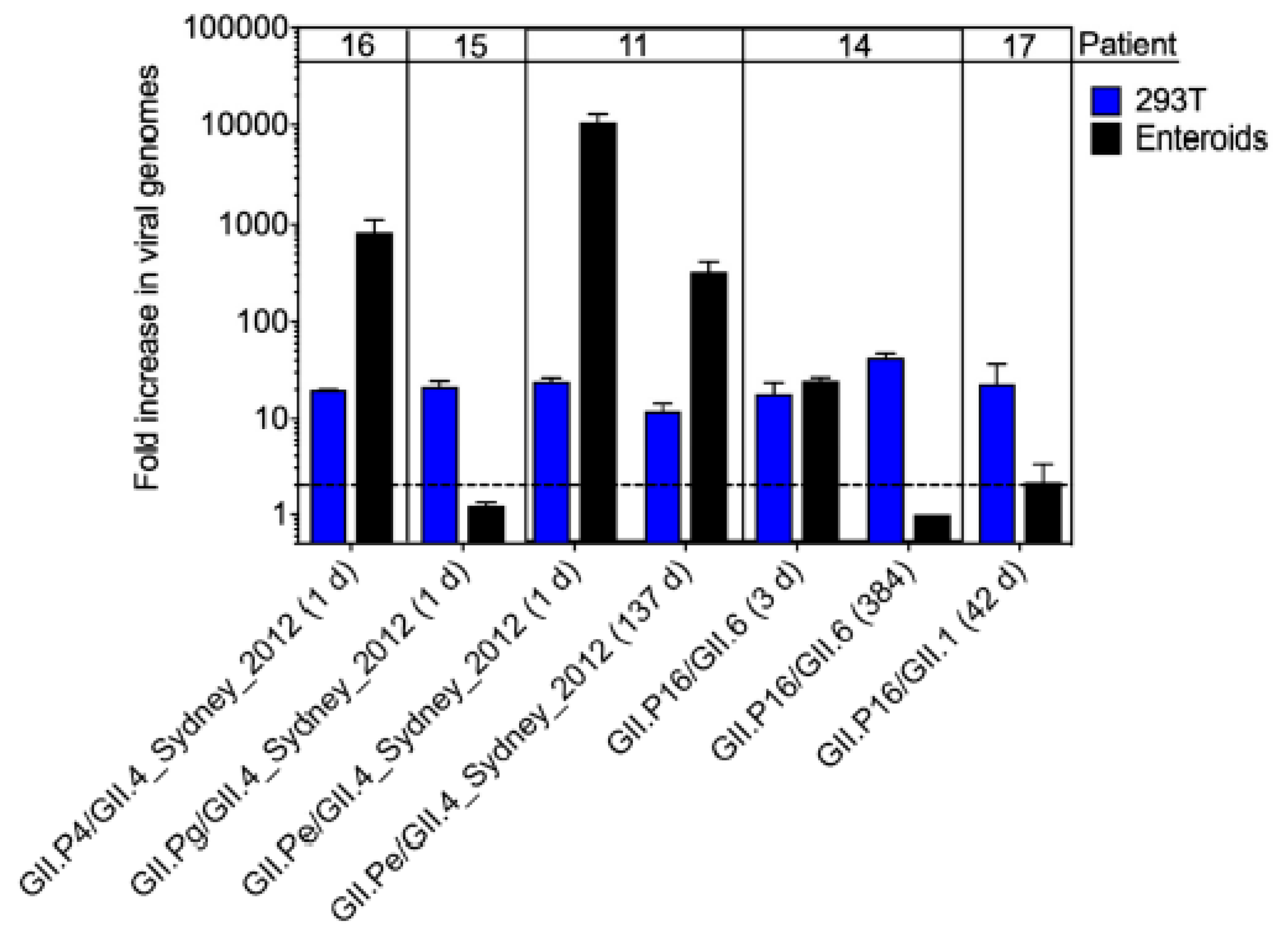
3.5. Infectivity of Noroviruses in Chronic Shedders
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marks, P.J.; Vipond, I.B.; Carlisle, D.; Deakin, D.; Fey, R.E.; Caul, E.O. Evidence for airborne transmission of Norwalk-like virus (NLV) in a hotel restaurant. Epidemiol. Infect. 2000, 124, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, L.; Hewitt, J.; Barclay, L.; Ahmed, S.M.; Lake, R.; Hall, A.J.; Lopman, B.; Kroneman, A.; Vennema, H.; Vinje, J.; et al. Norovirus genotype profiles associated with foodborne transmission, 1999–2012. Emerg. Infect. Dis. 2015, 21, 592–599. [Google Scholar] [CrossRef]
- Schorn, R.; Hohne, M.; Meerbach, A.; Bossart, W.; Wuthrich, R.P.; Schreier, E.; Muller, N.J.; Fehr, T. Chronic norovirus infection after kidney transplantation: Molecular evidence for immune-driven viral evolution. Clin. Infect. Dis. 2010, 51, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.M.; Widdowson, M.A.; Glass, R.I.; Akazawa, K.; Vinje, J.; Parashar, U.D. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.L.; Neill, F.H.; Estes, M.K.; Munoz, F.M.; Cameron, A.; DuPont, H.L.; Atmar, R.L. Noroviruses: The most common pediatric viral enteric pathogen at a large university hospital after introduction of rotavirus vaccination. J. Pediatric Infect. Dis. Soc. 2013, 2, 57–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bok, K.; Green, K.Y. Norovirus gastroenteritis in immunocompromised patients. N. Engl. J. Med. 2012, 367, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- Karst, S.M.; Baric, R.S. What is the reservoir of emergent human norovirus strains? J. Virol. 2015, 89, 5756–5759. [Google Scholar] [CrossRef] [Green Version]
- Lopman, B.; Vennema, H.; Kohli, E.; Pothier, P.; Sanchez, A.; Negredo, A.; Buesa, J.; Schreier, E.; Reacher, M.; Brown, D.; et al. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 2004, 363, 682–688. [Google Scholar] [CrossRef]
- Vinje, J.; Altena, S.A.; Koopmans, M.P. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 1997, 176, 1374–1378. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Donaldson, E.F.; Lobue, A.D.; Cannon, J.L.; Zheng, D.P.; Vinje, J.; Baric, R.S. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008, 5, e31. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Hansman, G.S.; Clancy, L.E.; Tanaka, M.M.; Rawlinson, W.D.; White, P.A. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 2005, 11, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Barclay, L.; Cannon, J.L.; Wikswo, M.E.; Phillips, A.R.; Browne, H.; Montmayeur, A.M.; Tatusov, R.L.; Burke, R.M.; Hall, A.J.; Vinje, J. Emerging Novel GII.P16 noroviruses associated with multiple capsid genotypes. Viruses 2019, 11, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, J.L.; Barclay, L.; Collins, N.R.; Wikswo, M.E.; Castro, C.J.; Magana, L.C.; Gregoricus, N.; Marine, R.L.; Chhabra, P.; Vinje, J. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of Novel GII.4 recombinant viruses. J. Clin. Microbiol. 2017, 55, 2208–2221. [Google Scholar] [CrossRef] [Green Version]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef]
- Green, K.Y. Norovirus infection in immunocompromised hosts. Clin. Microbiol. Infect. 2014, 20, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Frange, P.; Touzot, F.; Debre, M.; Heritier, S.; Leruez-Ville, M.; Cros, G.; Rouzioux, C.; Blanche, S.; Fischer, A.; Avettand-Fenoel, V. Prevalence and clinical impact of norovirus fecal shedding in children with inherited immune deficiencies. J. Infect. Dis. 2012, 206, 1269–1274. [Google Scholar] [CrossRef] [Green Version]
- Siebenga, J.J.; Beersma, M.F.; Vennema, H.; van Biezen, P.; Hartwig, N.J.; Koopmans, M. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: A model for in vivo molecular evolution. J. Infect. Dis. 2008, 198, 994–1001. [Google Scholar] [CrossRef] [Green Version]
- Chong, P.P.; Atmar, R.L. Norovirus in health care and implications for the immunocompromised host. Curr. Opin. Infect. Dis. 2019, 32, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Hakim, H.; Dallas, R.; Wolf, J.; Tang, L.; Schultz-Cherry, S.; Darling, V.; Johnson, C.; Karlsson, E.A.; Chang, T.C.; Jeha, S.; et al. Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clin. Infect. Dis. 2018, 67, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, T.; Singh, A.; Le Guyader, F.S.; La Rosa, G.; Saif, L.; McNeal, M. Multiplex real-time RT-PCR for the simultaneous detection and quantification of GI, GII and GIV noroviruses. J. Virol. Methods 2015, 223, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Vinje, J.; Hamidjaja, R.A.; Sobsey, M.D. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 2004, 116, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 2002, 100, 107–114. [Google Scholar] [CrossRef]
- Gao, Y.; Jin, M.; Cong, X.; Duan, Z.; Li, H.Y.; Guo, X.; Zuo, Y.; Zhang, Y.; Zhang, Y.; Wei, L. Clinical and molecular epidemiologic analyses of norovirus-associated sporadic gastroenteritis in adults from Beijing, China. J. Med. Virol. 2011, 83, 1078–1085. [Google Scholar] [CrossRef]
- Nishida, T.; Kimura, H.; Saitoh, M.; Shinohara, M.; Kato, M.; Fukuda, S.; Munemura, T.; Mikami, T.; Kawamoto, A.; Akiyama, M.; et al. Detection, quantitation, and phylogenetic analysis of noroviruses in Japanese oysters. Appl. Environ. Microbiol. 2003, 69, 5782–5786. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Cheon, D.S.; Kim, J.H.; Lee, D.H.; Jheong, W.H.; Heo, Y.J.; Chung, H.M.; Jee, Y.; Lee, J.S. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J. Clin. Microbiol. 2005, 43, 4836–4839. [Google Scholar] [CrossRef] [Green Version]
- Vennema, H.; de Bruin, E.; Koopmans, M. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 2002, 25, 233–235. [Google Scholar] [CrossRef]
- Boxman, I.L.; Tilburg, J.J.; Te Loeke, N.A.; Vennema, H.; Jonker, K.; de Boer, E.; Koopmans, M. Detection of noroviruses in shellfish in the Netherlands. Int. J. Food Microbiol. 2006, 108, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Kroneman, A.; Vennema, H.; Deforche, K.; Avoort, H.v.d.; Penarda, S.; Oberste, M.S.; Vinje, J.; Koopmans, M. An automated genotypg tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; de Graaf, M.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Human norovirus culture in B cells. Nat. Protoc. 2015, 10, 1939–1947. [Google Scholar] [CrossRef] [Green Version]
- Kolawole, A.O.; Mirabelli, C.; Hill, D.R.; Svoboda, S.A.; Janowski, A.B.; Passalacqua, K.D.; Rodriguez, B.N.; Dame, M.K.; Freiden, P.; Berger, R.P.; et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019, 15, e1008057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taube, S.; Kolawole, A.O.; Hohne, M.; Wilkinson, J.E.; Handley, S.A.; Perry, J.W.; Thackray, L.B.; Akkina, R.; Wobus, C.E. A mouse model for human norovirus. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.M.; Lopman, B.A.; Levy, K. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS ONE 2013, 8, e75922. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Burden of Norovirus Illness in the U.S. Available online: https://www.cdc.gov/norovirus/trends-outbreaks/burden-US.html (accessed on 15 November 2018).
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002, 16, 869–871. [Google Scholar] [CrossRef]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef]
- Costantini, V.; Morantz, E.K.; Browne, H.; Ettayebi, K.; Zeng, X.L.; Atmar, R.L.; Estes, M.K.; Vinje, J. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 2018, 24, 1453–1464. [Google Scholar] [CrossRef] [Green Version]
- Robles, J.D.; Cheuk, D.K.; Ha, S.Y.; Chiang, A.K.; Chan, G.C. Norovirus infection in pediatric hematopoietic stem cell transplantation recipients: Incidence, risk factors, and outcome. Biol. Blood Marrow Transplant. 2012, 18, 1883–1889. [Google Scholar] [CrossRef] [Green Version]
- Munir, N.; Liu, P.; Gastanaduy, P.; Montes, J.; Shane, A.; Moe, C. Norovirus infection in immunocompromised children and children with hospital-acquired acute gastroenteritis. J. Med. Virol. 2014, 86, 1203–1209. [Google Scholar] [CrossRef]
- Daniel-Wayman, S.; Fahle, G.; Palmore, T.; Green, K.Y.; Prevots, D.R. Norovirus, astrovirus, and sapovirus among immunocompromised patients at a tertiary care research hospital. Diagn. Microbiol. Infect. Dis. 2018, 92, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Escolano, A.L.; Velazquez, F.R.; Escobar-Herrera, J.; Lopez Saucedo, C.; Torres, J.; Estrada-Garcia, T. Human caliciviruses detected in Mexican children admitted to hospital during 1998–2000, with severe acute gastroenteritis not due to other enteropathogens. J. Med. Virol. 2010, 82, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, J.; de Graaf, M.; Smits, S.; Schapendonk, C.M.E.; Verjans, G.; Vennema, H.; van der Eijk, A.A.; Phan, M.V.T.; Cotten, M.; Koopmans, M. Whole-genome next-generation sequencing to study within-host evolution of norovirus (NoV) among immunocompromised patients with chronic NoV infection. J. Infect. Dis. 2017, 216, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Vega, E.; Donaldson, E.; Huynh, J.; Barclay, L.; Lopman, B.; Baric, R.; Chen, L.F.; Vinje, J. RNA populations in immunocompromised patients as reservoirs for novel norovirus variants. J. Virol. 2014, 88, 14184–14196. [Google Scholar] [CrossRef] [Green Version]
- Bok, K.; Prevots, D.R.; Binder, A.M.; Parra, G.I.; Strollo, S.; Fahle, G.A.; Behrle-Yardley, A.; Johnson, J.A.; Levenson, E.A.; Sosnovtsev, S.V.; et al. Epidemiology of norovirus infection among immunocompromised patients at a tertiary care research hospital, 2010–2013. Open Forum Infect. Dis. 2016, 3, ofw169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, S.L.; Bird, B.H.; Glass, G.E. Sex differences in immune responses and viral shedding following Seoul virus infection in Norway rats. Am. J. Trop. Med. Hyg. 2001, 65, 57–63. [Google Scholar] [CrossRef]
- Robinson, C.M.; Wang, Y.; Pfeiffer, J.K. Sex-dependent intestinal replication of an enteric virus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Sukhrie, F.H.; Siebenga, J.J.; Beersma, M.F.; Koopmans, M. Chronic shedders as reservoir for nosocomial transmission of norovirus. J. Clin. Microbiol. 2010, 48, 4303–4305. [Google Scholar] [CrossRef] [Green Version]


| Total Patients (n = 1140) | Infected Patients (n = 123) | ||||
|---|---|---|---|---|---|
| n | % | N | % | p-Value | |
| Sex | 0.893 | ||||
| Male | 646 | 57 | 69 | 56 | |
| Female | 494 | 43 | 54 | 44 | |
| Age (years) | 0.003 | ||||
| 0–5 | 463 | 41 | 69 | 56 | |
| 6–10 | 249 | 22 | 17 | 14 | |
| 11–15 | 214 | 19 | 22 | 18 | |
| 16–20 | 175 | 15 | 11 | 9 | |
| 21+ | 39 | 3 | 4 | 3 | |
| Race | 0.712 | ||||
| White | 822 | 72 | 95 | 77 | |
| Black | 222 | 19 | 18 | 15 | |
| Asian | 25 | 2 | 3 | 2 | |
| Multiple | 63 | 6 | 6 | 5 | |
| Other | 7 | 1 | 1 | 1 | |
| Unknown | 1 | 0 | 0 | - | |
| Diagnosis | 0.064 | ||||
| Hematologic Malignancy | 660 | 58 | 87 | 71 | |
| Solid Tumor | 273 | 24 | 21 | 17 | |
| Brain Tumor | 102 | 9 | 8 | 7 | |
| Hematologic Disorder | 52 | 5 | 4 | 3 | |
| Infectious Disease | 35 | 3 | 1 | 1 | |
| Primary Immunodeficiency | 18 | 2 | 2 | 2 | |
| Patient | Year | Days Shed | Age at Onset (Yrs) | Sex | Race | Diagnosis | BMT | GVHD | ANC # at Onset | ANC # during Infection | Fever at Onset | No. Time Points with Fever | Diarrhea at Onset | No. Time Points with Diarrhea | Vomiting at Onset | No. Time Points with Vomiting |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2012 | 51 | 2.9 | Female | Other | HM | N | - | Low | Low | N | 0/3 | Y | 3/3 | N | 1/3 |
| 2 | 2012 | 154 | 14.0 | Female | White | HM | N | - | Normal | Normal/Low | N | 4/8 | Y | 5/8 | Y | 5/8 |
| 3 | 2012 | 47 | 1.7 | Male | White | HM | Y | N | Low | Low | N | 0/2 | N | 0/2 | N | 0/2 |
| 4 | 2012 | 261 | 14.1 | Male | White | HM | N | - | Normal | Normal/Low | N | 0/6 | Y | 6/6 | N | 0/6 |
| 5 | 2012 | 71 | 6.8 | Female | White | HM | N | - | Normal | Low | N | 1/2 | N | 1/2 | N | 0/2 |
| 6 | 2013 | 203 | 2.3 | Male | White | HM | N | - | Low | Low | N | 1/3 | Y | 2/3 | N | 0/3 |
| 7 | 2013 | 50 | 15.6 | Female | Black | HM | Y | N | Low | Low | N | 0/2 | Y | 2/2 | Y | 1/2 |
| 8 | 2013 | 147 | 5.1 | Female | Black | HM | N | - | Normal | Normal | Y | 3/4 | N | 1/4 | N | 1/4 |
| 9 | 2013 | 83 | 1.9 | Female | White | HM | N | - | Low | Low/Normal | Y | 2/2 | N | 1/2 | N | 0/2 |
| 10 | 2013 | 225 | 2.1 | Male | White | HM | N | - | NR | Normal | N | 1/3 | Y | 3/3 | N | 0/3 |
| 11 | 2013 | 137 | 6.4 | Female | White | HM | N | - | Low | Low | Y | 2/2 | Y | 1/2 | N | 0/2 |
| 12 | 2014 | 118 | 11.6 | Male | White | HM | N | N | Low | Low | N | 3/11 | Y | 9/11 | Y | 8/11 |
| 13 | 2014 | 60 | 1.2 | Male | White | HM | N | - | Normal | NR | N | 0/1 + | N | 0/1 + | N | 0/1 + |
| 14 | 2014 | 418 | 1.0 | Male | White | HM | N | - | Low | Normal | N | 2/7 | Y | 6/7 | Y | 5/7 |
| 15 | 2015 | 66 | 13.0 | Female | White | HM | N | - | Low | Low | N | 1/3 | Y | 1/3 | Y | 1/3 |
| 16 | 2015 | 60 | 3.5 | Female | White | HM | N | - | Low | Low | N | 1/3 | Y | 2/3 | Y | 1/3 |
| 17 | 2015 | 37 | 1.8 | Female | White | HM | N | - | Normal | Low | N | 1/2 | N | 0/2 | N | 0/2 |
| 18 | 2016 | 99 | 10.1 | Male | White | ST | Y | Y | High | High/Normal | Y | 1/7 | N | 0/7 | N | 0/7 |
| 19 | 2016 | 77 | 3.6 | Female | White | HM | N | - | Low | Low | N | 0/2 | N | 0/2 | N | 0/2 |
| 20 | 2017 | 195 | 2.1 | Male | White | HM | Y | N | Low | Low/Normal | N | 2/5 | N | 1/5 | N | 0/5 |
| 17 | 2018 | 97 | 4.1 | Female | White | HM | Y | - | Low | Low/Normal | N | 1/14 | N | 11/14 | N | 2/14 |
| Patient | Infection Classification | New Infection | Cleared Infection | Coinfections Other Pathogens | Genotype (ORF1/ORF2) | Infectious Virus Detected |
|---|---|---|---|---|---|---|
| 1 | Coinfection | Unknown | Unknown | None | /GI.5 and GII.P4/GII.4 Den Haag 2006b | Y |
| 2 | Single | Y | Y | C. difficile, astrovirus | GII.Pg/GII.6 | Y |
| 3 | Sequential & | Unknown | Y | C. difficile | GI.P3/GI.3 | Y |
| 4 | Single | Unknown | Unknown | C. difficile | GII.P16/GII.3 | Y |
| 5 | Single | Unknown | Unknown | Adenovirus | GII.Pe/GII.1 | Y |
| 6 | Single | Y | Unknown | Rotavirus | GII.Pe/GII.4 Sydney 2012 | Y |
| 7 | Single | Y | Y | CMV, BK Virus | GII.Pg/GII.4 Sydney 2012 | N |
| 8 | Single | Unknown | Y | None | /GII.4 Sydney 2012 | Y |
| 9 | Single | Y | Unknown | C. difficile | GII.P16/GII.2 | Y |
| 10 | Single | Y | Unknown | C. difficile, Epstein-Barr virus | GII.Pe/GII.4 Sydney 2012 | Y |
| 11 | Single | Unknown | Unknown | None | GII.Pe/GII.4 Sydney 2012 | Y |
| 12 | Single | Y | Unknown | K. pneumoniae, S. epidermidis, BK virus, adenovirus, parainfluenza virus | GII.P4 New Orleans 2009/GII.4 Sydney 2012 | Y |
| 13 | Single | Y | Unknown | None | GI.P3/GI.3 | Y |
| 14 | Single | Y | Unknown | Adenovirus, parainfluenza virus | GII.P16/GII.6 | Y |
| 15 | Sequential $ | Unknown | Y | C. difficile | GII.Pg/GII.4 Sydney 2012 | Y |
| 16 | Superinfection * | Y | Y | Human rhinovirus, enterovirus | GII.P4/GII.4 Sydney 2012 | Y |
| 17 | Sequential # | Y | Y | Human rhinovirus, enterovirus | /GII.6 | Y |
| 18 | Sequential @ | Y | Unknown | C. parapsilosis, adenovirus | GII.Pg/GII.4 Sydney 2012 | Y |
| 19 | Single | Y | Unknown | None | GII.Pe/GII.4 Sydney 2012 | Y |
| 20 | Single | Y | Unknown | C. difficile, astrovirus | GII.P16/GII.4 Sydney 2012 | Y |
| 17 | Sequential ^ | Y | Unknown | None | GII.Pg and GII.P16/GII.1 | Y |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davis, A.; Cortez, V.; Grodzki, M.; Dallas, R.; Ferrolino, J.; Freiden, P.; Maron, G.; Hakim, H.; Hayden, R.T.; Tang, L.; et al. Infectious Norovirus Is Chronically Shed by Immunocompromised Pediatric Hosts. Viruses 2020, 12, 619. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060619
Davis A, Cortez V, Grodzki M, Dallas R, Ferrolino J, Freiden P, Maron G, Hakim H, Hayden RT, Tang L, et al. Infectious Norovirus Is Chronically Shed by Immunocompromised Pediatric Hosts. Viruses. 2020; 12(6):619. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060619
Chicago/Turabian StyleDavis, Amy, Valerie Cortez, Marco Grodzki, Ronald Dallas, Jose Ferrolino, Pamela Freiden, Gabriela Maron, Hana Hakim, Randall T. Hayden, Li Tang, and et al. 2020. "Infectious Norovirus Is Chronically Shed by Immunocompromised Pediatric Hosts" Viruses 12, no. 6: 619. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060619