Arbuscular Mycorrhizal Symbiosis Primes Tolerance to Cucumber Mosaic Virus in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials, AMF Inoculation, and Virus Infection
2.2. Evaluation of Mycorrhizal Colonization, CMV Infection, and Viral Replication
2.3. Illumina Sequencing, Bioinformatic Analysis, and qRT-PCR Validation
2.4. Leaf Gas Exchange Measurements
2.5. Hormone Measurements
3. Results
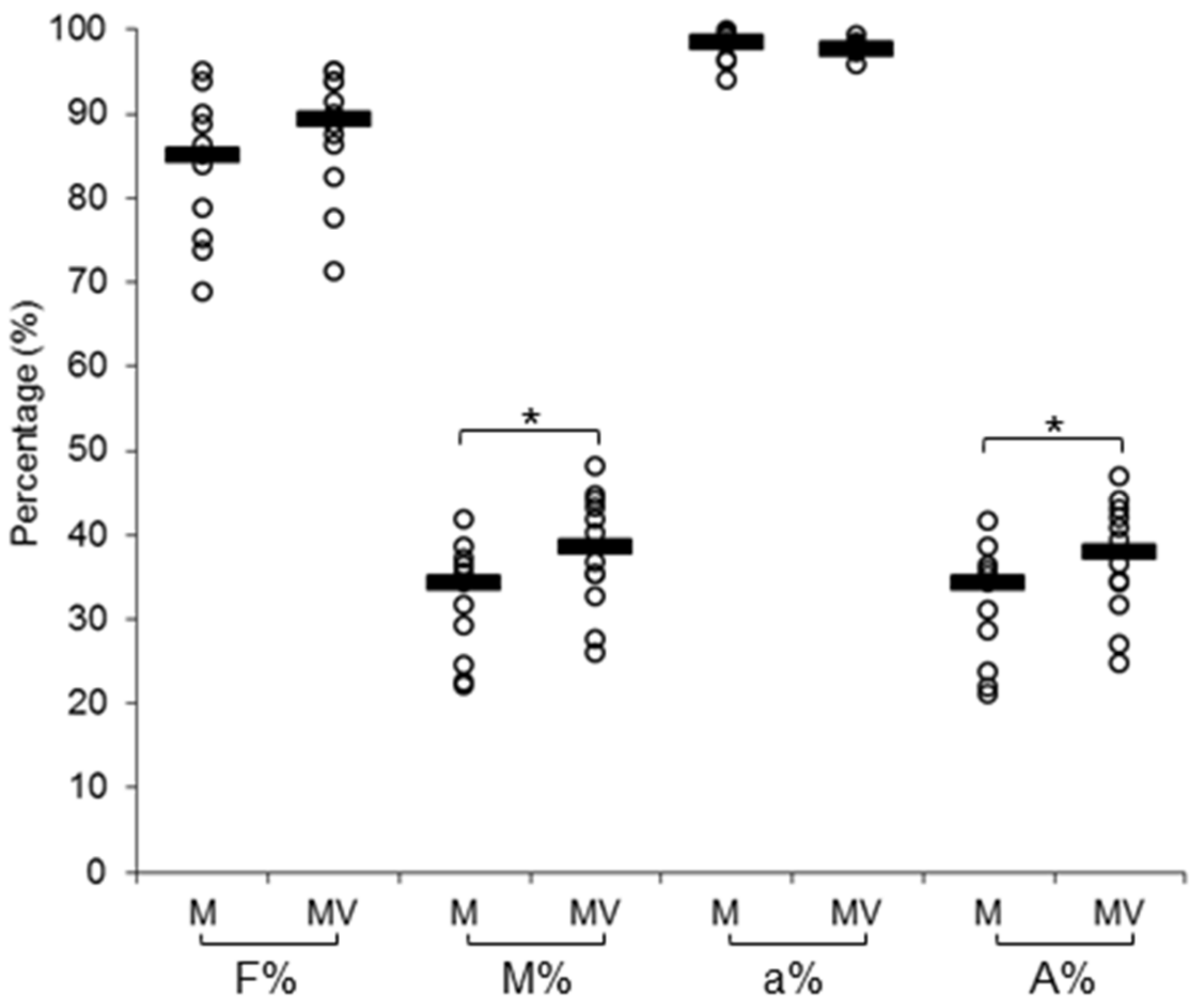
3.1. Mycorrhizal Colonization and Viral Infection
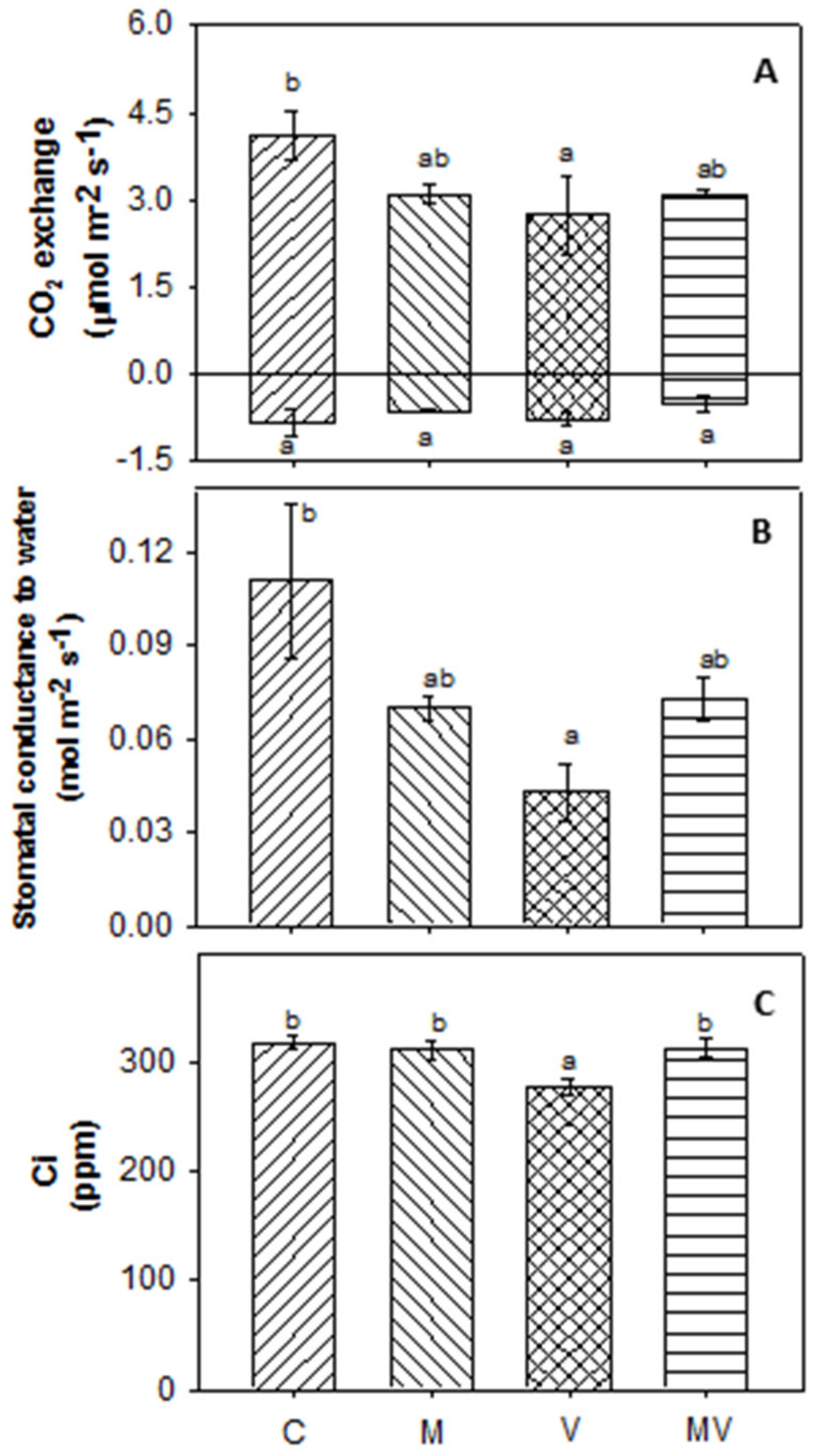
3.2. Biomass Production and Allocation and Leaf Gas Exchange
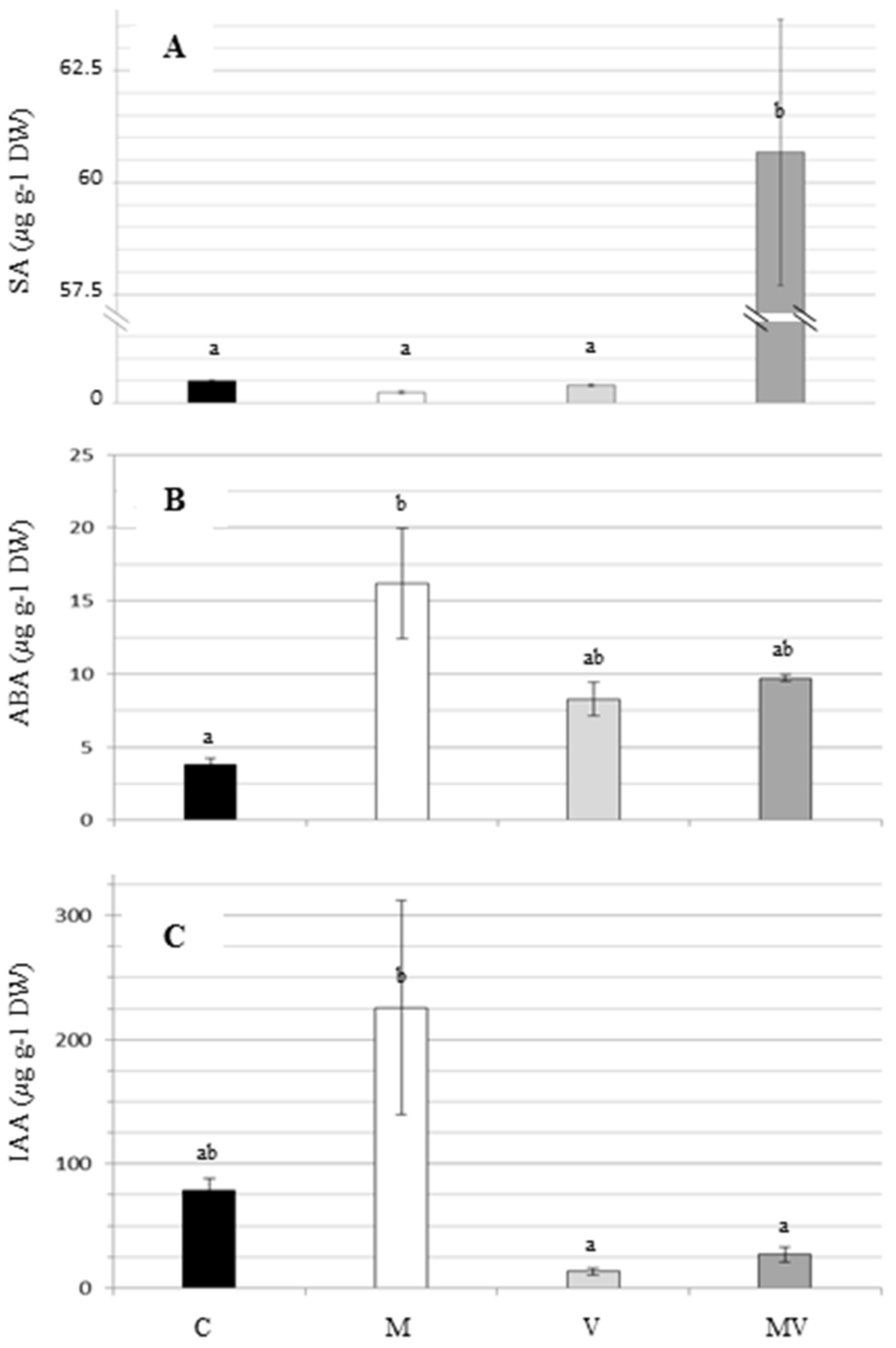
3.3. Hormones Measurement
3.4. RNA-seq Data and Functional Characterization of DEGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
Abbreviations
| CMV | cucumber mosaic virus |
| AMF | arbuscular mycorrhizal fungi |
| MIR | mycorrhiza-induced resistance |
| MIS | mycorrhiza-induced susceptibility |
| GO | gene ontology |
| ROS | reactive oxygen species |
| qRT-PCR | quantitative RT-PCR |
| Dpi | days post inoculation |
| C | control plants |
| V | virus-inoculated plants |
| M | mycorrhizal plants |
| MV | mycorrhizal virus-inoculated plants |
| JA | jasmonic acid |
| SA | salicylic acid |
| ABA | abscisic acid |
| IAA | indolacetic acid |
| F% | mycorrhization frequency |
| M% | mycorrhization intensity |
| a% | arbuscules in the colonized segments of roots |
| A% | arbuscules in the whole root |
| DSI | disease severity index |
| Ci | substomatal CO2 concentration |
| DEGs | differentially expressed genes |
References
- Hanssen, I.; Lapidot, M.; Thomma, B. Emerging Viral Diseases of Tomato Crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybicki, E. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Sevik, M.; Arli-Sokmen, M. Estimation of the effect of Tomato spotted wilt virus (TSWV) infection on some yield components of tomato. Phytoparasitica 2012, 40, 87–93. [Google Scholar] [CrossRef]
- Strange, R.; Scott, P. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Nogués, S.; Alegre, L. An increase in water deficit has no impact on the photosynthetic capacity of field-grown Mediterranean plants. Funct. Plant Biol. 2002, 29, 10. [Google Scholar] [CrossRef]
- Culver, J.; Padmanabhan, M. Virus-induced disease: Altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 2007, 45, 221–243. [Google Scholar] [CrossRef] [Green Version]
- Davino, S.; Miozzi, L.; Panno, S.; Rubio, L.; Davino, M.; Accotto, G. Recombination profiles between Tomato yellow leaf curl virus and Tomato yellow leaf curl Sardinia virus in laboratory and field conditions: Evolutionary and taxonomic implications. J. Gen. Virol. 2012, 93, 2712–2717. [Google Scholar] [CrossRef] [Green Version]
- Duffy, S.; Holmes, E. Validation of high rates of nucleotide substitution in geminiviruses: Phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 2009, 90, 1539–1547. [Google Scholar] [CrossRef]
- Lauring, A.; Frydman, J.; Andino, R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 2013, 11, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef] [Green Version]
- Cahill, M.; Gorman, K.; Day, S.; Denholm, I.; Elbert, A.; Nauen, R. Baseline determination and detection of resistance to imidacloprid in Bemisia tabaci (Homoptera: Aleyrodidae). Bull. Entomol. Res. 1996, 86, 343–349. [Google Scholar] [CrossRef]
- Cahill, M.; Jarvis, W.; Gorman, K.; Denholm, I. Resolution of baseline responses and documentation of resistance to buprofezin in Bemisia tabaci (Homoptera: Aleyrodidae). Bull. Entomol. Res. 1996, 86, 117–122. [Google Scholar] [CrossRef]
- Castle, S.; Palumbo, J.; Prabhaker, N. Newer insecticides for plant virus disease management. Virus Res. 2009, 141, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Mochizuki, T.; Ohki, S.T. Cucumber mosaic virus: Viral genes as virulence determinants. Mol. Plant Pathol. 2012, 13, 217–225. [Google Scholar] [CrossRef]
- Lanfranco, L.; Bonfante, P.; Genre, A. The Mutualistic Interaction between Plants and Arbuscular Mycorrhizal Fungi. Microbiol Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Gianinazzi, S.; Gollotte, A.; Binet, M.; van Tuinen, D.; Redecker, D.; Wipf, D. Agroecology: The key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 2010, 20, 519–530. [Google Scholar] [CrossRef]
- Johnson, N.C.; Wilson, G.W.T.; Wilson, J.A.; Miller, R.M.; Bowker, M.A. Mycorrhizal phenotypes and the Law of the Minimum. New Phytol. 2015, 205, 1473–1484. [Google Scholar] [CrossRef]
- Vannette, R.; Hunter, M. Mycorrhizal fungi as mediators of defence against insect pests in agricultural systems. Agric. For. Entomol. 2009, 11, 351–358. [Google Scholar] [CrossRef]
- Chitarra, W.; Pagliarani, C.; Maserti, B.; Lumini, E.; Siciliano, I.; Cascone, P.; Schubert, A.; Gambino, G.; Balestrini, R.; Guerrieri, E. Insights on the Impact of Arbuscular Mycorrhizal Symbiosis on Tomato Tolerance to Water Stress. Plant Physiol. 2016, 171, 1009–1023. [Google Scholar] [CrossRef] [Green Version]
- Rivero, J.; Álvarez, D.; Flors, V.; Azcón-Aguilar, C.; Pozo, M.J. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol. 2018, 220, 1322–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreño, Á.; Molina, S.; Andreo-Jiménez, B.; Porcel, R.; García-Mina, J.M.; Ruyter-Spira, C.; López-Ráez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Env. 2016, 39, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Volpe, V.; Chitarra, W.; Cascone, P.; Volpe, M.G.; Bartolini, P.; Moneti, G.; Pieraccini, G.; Di Serio, C.; Maserti, B.; Guerrieri, E.; et al. The Association With Two Different Arbuscular Mycorrhizal Fungi Differently Affects Water Stress Tolerance in Tomato. Front Plant Sci. 2018, 9, 1480. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Martinez-Medina, A.; Lopez-Raez, J.; Pozo, M. Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.; Pozo, M.; Ton, J.; van Dam, N.; Conrath, U. Recognizing Plant Defense Priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, D.D.; Neal, A.L.; van Wees, S.C.; Ton, J. Mycorrhiza-induced resistance: More than the sum of its parts? Trends Plant Sci. 2013, 18, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Pozo, M.J.; Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 2007, 10, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Fiorilli, V.; Catoni, M.; Miozzi, L.; Novero, M.; Accotto, G.P.; Lanfranco, L. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 2009, 184, 975–987. [Google Scholar] [CrossRef]
- López-Ráez, J.A.; Charnikhova, T.; Fernández, I.; Bouwmeester, H.; Pozo, M.J. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J. Plant Physiol. 2011, 168, 294–297. [Google Scholar] [CrossRef]
- Wright, D.; Read, D.; Scholes, J. Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ. 1998, 21, 881–891. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Aroca, R. Host response to osmotic stresses: Stomatal behaviour and water use efficiency of arbuscular mycorrhizal plants. In Arbuscular Mycorrhizas: Physiology and Function; Springer: Dordrecht, Germany, 2010; pp. 239–256. [Google Scholar]
- López-Ráez, J.A.; Flors, V.; García, J.M.; Pozo, M.J. AM symbiosis alters phenolic acid content in tomato roots. Plant Signal Behav. 2010, 5, 1138–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Ráez, J.A.; Verhage, A.; Fernández, I.; García, J.M.; Azcón-Aguilar, C.; Flors, V.; Pozo, M.J. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 2010, 61, 2589–2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miozzi, L.; Catoni, M.; Fiorilli, V.; Mullineaux, P.M.; Accotto, G.P.; Lanfranco, L. Arbuscular mycorrhizal symbiosis limits foliar transcriptional responses to viral infection and favors long-term virus accumulation. Mol. Plant Microbe Interact 2011, 24, 1562–1572. [Google Scholar] [CrossRef] [Green Version]
- Miozzi, L.; Vaira, A.M.; Catoni, M.; Fiorilli, V.; Accotto, G.P.; Lanfranco, L. Arbuscular Mycorrhizal Symbiosis: Plant Friend or Foe in the Fight against Viruses? Front Microbiol. 2019, 10, 1238. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, E. Sand and water culture methods used in the study of plant nutrition, Commonwealth Bureau of Horticultural and Plantation Crops, East Mailing. Tech. Commun. 1966, 22. [Google Scholar] [CrossRef]
- Maffei, G.; Miozzi, L.; Fiorilli, V.; Novero, M.; Lanfranco, L.; Accotto, G.P. The arbuscular mycorrhizal symbiosis attenuates symptom severity and reduces virus concentration in tomato infected by Tomato yellow leaf curl Sardinia virus (TYLCSV). Mycorrhiza 2014, 24, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P.; Roossinck, M.; Dietzgen, R.; Francki, R. Cucumber Mosaic Virus. Adv. Virus Res. 1992, 41, 281–348. [Google Scholar]
- Trouvelot, A.; Kough, J.; Gianinazzi-Pearson, V. Estimation of VA mycorrhizal infection levels. Research for methods having a functional significance. In Physiological and Genetical Aspects of Mycorrhizae Aspects Physiologiques et Genetiques des Mycorhizes, Dijon (France), 1-5 Jul 1985. /1INRA; INRA Presse: Paris, France, 1986. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.; Sauvageau, M.; Goff, L.; Rinn, J.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46. [Google Scholar] [CrossRef]
- Burset, M.; Guigo, R. Evaluation of gene structure prediction programs. Genomics 1996, 34, 353–367. [Google Scholar] [CrossRef]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruger, P.; Selbig, J.; Muller, L.; Rhee, S.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Chitarra, W.; Perrone, I.; Avanzato, C.G.; Minio, A.; Boccacci, P.; Santini, D.; Gilardi, G.; Siciliano, I.; Gullino, M.L.; Delledonne, M.; et al. Grapevine Grafting: Scion Transcript Profiling and Defense-Related Metabolites Induced by Rootstocks. Front Plant Sci. 2017, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.F.; Reddy, M.S.; Ryu, C.M.; Kloepper, J.W.; Li, R. Rhizobacteria-Mediated Growth Promotion of Tomato Leads to Protection Against Cucumber mosaic virus. Phytopathology 2003, 93, 1301–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P. Plant hormone-mediated regulation of stress responses. BMC Plant Biology 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.H. Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.P.; Liu, D.S.; Yan, T.; Fang, X.D.; Dong, K.; Xu, J.; Wang, Y.; Yu, J.L.; Wang, X.B. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog 2017, 13, e1006522. [Google Scholar] [CrossRef] [Green Version]
- Nemes, K.; Gellert, A.; Boka, K.; Vagi, P.; Salanki, K. Symptom recovery is affected by Cucumber mosaic virus coat protein phosphorylation. Virology 2019, 536, 68–77. [Google Scholar] [CrossRef]
- Jovel, J.; Walker, M.; Sanfacon, H. Recovery of Nicotiana benthamiana plants from a necrotic response induced by a nepovirus is associated with RNA silencing but not with reduced virus titer. J. Virol. 2007, 81, 12285–12297. [Google Scholar] [CrossRef] [Green Version]
- Ghoshal, B.; Sanfacon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Shaul, O.; Galili, S.; Volpin, H.; Ginzberg, I.; Elad, Y.; Chet, I.; Kapulnik, Y. Mycorrhiza-induced changes in disease severity and PR protein expression in tobacco leaves. Mol. Plant Microbe Interact 1999, 12, 1000–1007. [Google Scholar] [CrossRef] [Green Version]
- Sipahioglu, M.H.; Demir, S.; Usta, M.; Akkopru, A. Biological relationship of Potato Virus Y and arbuscular mycorrhizal fangus Glomus intraradices in potato. Pest Technol. 2009, 3, 4. [Google Scholar]
- Gamir, J.; Sánchez-Bel, P.; Flors, V. Molecular and physiological stages of priming: How plants prepare for environmental challenges. Plant Cell Rep. 2014, 33, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 174–189. [Google Scholar]
- Pozo, M.J.; López-Ráez, J.A.; Azcón-Aguilar, C.; García-Garrido, J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 2015, 28, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic Acid as Pathogen Effector and Immune Regulator. Front Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense Priming: An Adaptive Part of Induced Resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [Green Version]
- Miransari, M.; Abrishamchi, A.; Khoshbakht, K.; Niknam, V. Plant hormones as signals in arbuscular mycorrhizal symbiosis. Crit. Rev. Biotechnol. 2014, 34, 123–133. [Google Scholar] [CrossRef]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Petti, C.; Reiber, K.; Ali, S.S.; Berney, M.; Doohan, F.M. Auxin as a player in the biocontrol of Fusarium head blight disease of barley and its potential as a disease control agent. BMC Plant Biol. 2012, 12, 224. [Google Scholar] [CrossRef] [Green Version]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Camejo, D.; Guzmán-Cedeño, Á.; Moreno, A. Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Riedle-Bauer, M. Role of Reactive Oxygen Species and Antioxidant Enzymes in Systemic Virus Infections of Plants. J. Phytopathol. 2000, 148, 297–302. [Google Scholar] [CrossRef]
- Cervantes-Gámez, R.G.; Bueno-Ibarra, M.A.; Cruz-Mendívil, A.; Calderón-Vázquez, C.L.; Ramírez-Douriet, C.M.; Maldonado-Mendoza, I.E.; Villalobos-López, M.Á.; Valdez-Ortíz, Á.; López-Meyer, M. Arbuscular mycorrhizal symbiosis-induced expression changes in Solanum lycopersicum leaves revealed by RNA-seq analysis. Plant Mol. Biol. Report. 2016, 34, 89–102. [Google Scholar] [CrossRef]
- Mathur, S.; Sharma, M.P.; Jajoo, A. Improved photosynthetic efficacy of maize (Zea mays) plants with arbuscular mycorrhizal fungi (AMF) under high temperature stress. J. Photochem. Photobiol. B 2018, 180, 149–154. [Google Scholar] [CrossRef]
- Aseel, D.G.; Rashad, Y.M.; Hammad, S.M. Arbuscular Mycorrhizal Fungi Trigger Transcriptional Expression of Flavonoid and Chlorogenic Acid Biosynthetic Pathways Genes in Tomato against Tomato Mosaic Virus. Sci. Rep. 2019, 9, 9692. [Google Scholar] [CrossRef] [Green Version]
- Zouari, I.; Salvioli, A.; Chialva, M.; Novero, M.; Miozzi, L.; Tenore, G.C.; Bagnaresi, P.; Bonfante, P. From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genom. 2014, 15, 221. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, D.; Partridge, J.; Roberts, N.; Boonham, N.; Foster, G. The Effects of Plant Virus Infection on Polarization Reflection from Leaves. PLoS ONE 2016, 11, e0152836. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miozzi, L.; Vaira, A.M.; Brilli, F.; Casarin, V.; Berti, M.; Ferrandino, A.; Nerva, L.; Accotto, G.P.; Lanfranco, L. Arbuscular Mycorrhizal Symbiosis Primes Tolerance to Cucumber Mosaic Virus in Tomato. Viruses 2020, 12, 675. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060675
Miozzi L, Vaira AM, Brilli F, Casarin V, Berti M, Ferrandino A, Nerva L, Accotto GP, Lanfranco L. Arbuscular Mycorrhizal Symbiosis Primes Tolerance to Cucumber Mosaic Virus in Tomato. Viruses. 2020; 12(6):675. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060675
Chicago/Turabian StyleMiozzi, Laura, Anna Maria Vaira, Federico Brilli, Valerio Casarin, Mara Berti, Alessandra Ferrandino, Luca Nerva, Gian Paolo Accotto, and Luisa Lanfranco. 2020. "Arbuscular Mycorrhizal Symbiosis Primes Tolerance to Cucumber Mosaic Virus in Tomato" Viruses 12, no. 6: 675. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060675






