Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow™ and qRT-PCR Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics and Samples Collection
2.2. Reagents
2.3. Virus and Cells
2.4. Human PBMC Culture and OROV Infection
2.5. Whole Blood Infection
2.6. RNA Extraction and qRT-PCR for OROV Quantitation
2.7. qRT-PCR for Gene Expression
2.8. Focus Forming Assay (FFA)
2.9. Single-Cell OROV Detection by RNA PrimeFlow™
2.10. OROV Proteins, Genome and Antigenome Detection by Confocal Microscopy
2.11. Immunofluorescence and Flow Cytometry
2.12. Graphics and Statistical Analysis
3. Results
3.1. Detection of OROV Genome and Antigenome by RNA Primeflow™ Assay
3.2. OROV Productively Infects Different Human Leukocytes Lineages
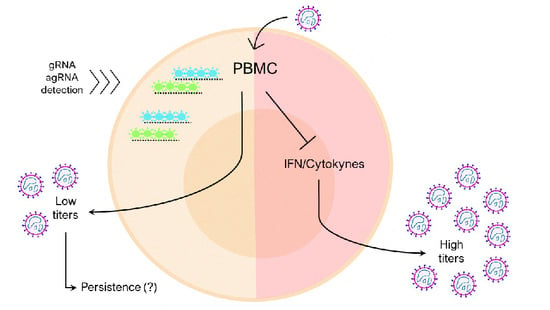
3.3. Human Peripheral Blood Leukocytes Are Susceptible to OROV Infection but Do Not Generate Large Quantities of Infectious Particles
3.4. Human PBMCs Response to OROV Infection Involves Upregulation of IFNs and ISGs Expression
3.5. IFNAR Blocking and Glucocorticoid Treatment Favor OROV Replication in PBMCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mavalankar, D.; Shastri, P.; Bandyopadhyay, T.; Parmar, J.; Ramani, K.V. Increased mortality rate associated with Chikungunya epidemic, Ahmedabad, India. Emerg. Infect. Dis. 2008, 14, 412–415. [Google Scholar] [CrossRef]
- Travassos Da Rosa, J.F.; De Souza, W.M.; De Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Teixeira Nunes, M.R. Oropouche virus: Clinical, epidemiological, and molecular aspects of a neglected orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- de Souza Luna, L.K.; Rodrigues, A.H.; Santos, R.I.M.; Sesti-Costa, R.; Criado, M.F.; Martins, R.B.; Silva, M.L.; Delcaro, L.S.; Proença-Modena, J.L.; Figueiredo, L.T.M.; et al. Oropouche Virus Is Detected in Peripheral Blood Leukocytes From Patients. J. Med. Virol. 2017, 89, 1108–1111. [Google Scholar] [CrossRef]
- Cardoso, B.F.; Serra, O.P.; Da Silva Heinen, L.B.; Zuchi, N.; De Souza, V.C.; Naveca, F.G.; Dos Santos, M.A.M.; Slhessarenko, R.D. Detection of oropouche virus segment s in patients and in culex quinquefasciatus in the state of mato grosso, Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 745–754. [Google Scholar] [CrossRef]
- Vasconcelos, H.B.; Azevedo, R.S.S.; Casseb, S.M.; Nunes-neto, J.P.; Chiang, J.O.; Cantuária, P.C.; Segura, M.N.O.; Martins, L.C.; Monteiro, H.A.O.; Rodrigues, S.G.; et al. Oropouche fever epidemic in Northern Brazil: Epidemiology and molecular characterization of isolates. J. Clin. Virol. 2008, 44, 129–133. [Google Scholar] [CrossRef]
- Teixeira Nunes, M.R.; Caricio Martins, L.; Guerreiro Rodrigues, S.; Oliveira Chiang, J.; da Silva Azevedo, R.D.S.; Da Rosa, A.P.T.; da Costa Vasconcelos, P.F. Oropouche Virus Isolation, Southeast Brazil. Emerg. Infect. Dis. 2005, 11, 1610–1613. [Google Scholar] [CrossRef]
- Bastos, M.D.S.; Figueiredo, L.T.M.; Naveca, F.G.; Monte, R.L.; Lessa, N.; De Figueiredo, R.M.P.; Gimaque, J.B.D.L.; João, G.P.; Ramasawmy, R.; Mourão, M.P.G. Short report: Identification of Oropouche Orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am. J. Trop. Med. Hyg. 2012, 86, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Romero-Alvarez, D.; Escobar, L.E. Oropouche fever, an emergent disease from the Americas. Microbes Infect. 2018, 20, 135–146. [Google Scholar] [CrossRef]
- Mourão, M.P.G.; Bastos, M.S.; Gimaque, J.B.L.; Mota, B.R.; Souza, G.S.; Grimmer, G.H.N.; Galusso, E.S.; Arruda, E.; Figueiredo, L.T.M. Oropouche fever outbreak, Manaus, Brazil, 2007-2008. Emerg. Infect. Dis. 2009, 15, 2063–2064. [Google Scholar] [CrossRef]
- Liang, G.; Gao, X.; Gould, E.A. Factors responsible for the emergence of arboviruses; strategies, challenges and limitations for their control. Emerg. Microbes Infect. 2015, 4, 1–5. [Google Scholar] [CrossRef]
- Elliott, R.M. Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 2014, 12, 673–685. [Google Scholar] [CrossRef]
- Tilston-Lunel, N.L.; Acrani, G.O.; Randall, R.E.; Elliott, R.M. Generation of Recombinant Oropouche Viruses Lacking the Nonstructural Protein NSm or NSs. J. Virol. 2016, 90. [Google Scholar] [CrossRef] [Green Version]
- Haller, O.; Kochs, G.; Weber, F. The interferon response circuit: Induction and suppression by pathogenic viruses. Virology 2006, 344, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.K.; Sharpe, A.H. Dendritic cells giveth and taketh away. Nat. Immunol. 2005, 6, 227–228. [Google Scholar] [CrossRef]
- Mellman, I. Dendritic cells: Master regulators of the immune response. Cancer Immunol. Res. 2013, 1, 145–149. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Hua, S.; Chen, H.; Ouyang, Z.; Einkauf, K.; Tse, S.; Ard, K.; Ciaranello, A.; Yawetz, S.; Sax, P.; et al. Transcriptional changes during naturally-acquired Zika Virus infection render dendritic cells highly conducive to viral replication. Cell Rep. 2017, 21, 3471–3482. [Google Scholar] [CrossRef] [Green Version]
- Foo, S.; Chen, W.; Chan, Y.; Bowman, J.W.; Chang, L.; Choi, Y.; Yoo, J.S.; Ge, J.; Cheng, G.; Bonnin, A.; et al. Asian Zika virus strains target CD14+ blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat. Microbiol. 2017. [CrossRef] [Green Version]
- Proenca-Modena, J.L.; Sesti-Costa, R.; Pinto, A.K.; Richner, J.M.; Lazear, H.M.; Lucas, T.; Hyde, J.L.; Diamond, M.S. Oropouche virus infection and pathogenesis are restricted by MAVS, IRF-3, IRF-7, and type I interferon signaling pathways in nonmyeloid cells. J. Virol. 2015, 89, 4720–4737. [Google Scholar] [CrossRef] [Green Version]
- O’Doherty, U.; Swiggard, W.J.; Malim, M.H. Human Immunodeficiency Virus Type 1 Spinoculation Enhances Infection through Virus Binding. J. Virol. 2000, 74, 10074–10080. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Ma, J.; Li, J.; Li, D.; Li, G.; Li, F.; Zhang, Q.; Yu, H.; Yasui, F.; Ye, C.; et al. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J. Clin. Invest. 2017, 127, 269–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Almeida, A.R.; Dantas, A.T.; Pereira, M.C.; Cordeiro, M.F.; Gonçalves, R.S.G.; de Melo Rêgo, M.J.B.; da Rocha Pitta, I.; Duarte, A.L.B.P.; da Rocha Pitta, M.G. Dexamethasone inhibits cytokine production in PBMC from systemic sclerosis patients. Inflammopharmacology 2019, 27, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Schöbel, A.; Rösch, K.; Herker, E. Functional innate immunity restricts Hepatitis C Virus infection in induced pluripotent stem cell-derived hepatocytes. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, W.; Earley, L.F.; Chai, Z.; Chen, X.; Sun, J.; He, T.; Deng, M.; Hirsch, M.L.; Ting, J.; Samulski, R.J.; et al. Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zhou, L.; Xie, H.; Zheng, S. Innate immune evasion by hepatitis B virus-mediated downregulation of TRIF. Biochem. Biophys. Res. Commun. 2015, 463, 719–725. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Zhu, J.; Sarkar, S.N.; Coyne, C.B. Regulation of mitochondrial antiviral signaling (MAVS) expression and signaling by the mitochondria-associated endoplasmic reticulum membrane (MAM) protein Gp78. J. Biol. Chem. 2014, 289, 1604–1616. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Zhang, H.; Qiu, J.; Lin, Y.; Liang, J.; Xiao, X.; Fu, L.; Wang, F.; Cai, J.; Tan, Y.; et al. Activation of cyclic adenosine monophosphate pathway increases the sensitivity of cancer cells to the oncolytic Virus M1. Mol. Ther. 2016, 24, 156–165. [Google Scholar] [CrossRef]
- Harper, M.S.; Guo, K.; Gibbert, K.; Lee, E.J.; Dillon, S.M.; Barrett, B.S.; Mccarter, M.D.; Hasenkrug, K.J.; Dittmer, U. Interferon- α Subtypes in an Ex Vivo Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms. PLoS Pathog. 2015, 11, e10052541. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, W.; Duggan, E.S.; Booth, J.L.; Zou, M.-H.; Metcalf, J.P. HHS Public Access. Virology 2015, 482, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Anafu, A.A.; Bowen, C.H.; Chin, C.R.; Brass, A.L.; Holm, G.H. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013, 288, 17261–17271. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Kim, K.W.; Joo, K.; Kim, J.C. Angiogenin ameliorates corneal opacity and neovascularization via regulating immune response in corneal fibroblasts. BMC Ophthalmol. 2016, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Micke, P.; Ohshima, M.; Tahmasebpoor, S.; Ren, Z.P.; Östman, A.; Pontén, F.; Botling, J. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab. Investig. 2006, 86, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Zwicker, S.; Bureik, D.; Bosma, M.; Martinez, G.L.; Almer, S.; Boström, E.A. Receptor-type protein-tyrosine phosphatase ζ and colony stimulating factor-1 receptor in the intestine: Cellular expression and cytokine-and chemokine responses by interleukin-34 and colony stimulating factor-1. PLoS ONE 2016, 11, e0167324. [Google Scholar] [CrossRef]
- Bowen, J.R.; Quicke, K.M.; Maddur, M.S.; Neal, J.T.O.; Mcdonald, E.; Fedorova, N.B.; Puri, V.; Shabman, R.S.; Pulendran, B.; Suthar, M.S. Zika Virus Antagonizes Type I Interferon Responses during Infection of Human Dendritic Cells. PLoS Pathog. 2017, 13, e1006164. [Google Scholar] [CrossRef]
- Tsang, M.; Gantchev, J.; Ghazawi, F.M.; Litvinov, I.V. Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture. Biotechniques 2017, 63, 230–233. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, N.S.; Mendonça, L.R.; Dias, M.V.S.; Pontelli, M.C.; da Silva, Z.M.; Criado, M.F.; da Silva-Januário, M.E.; Schindler, M.; Jamur, M.C.; Oliver, C.; et al. ESCRT machinery components are required for Orthobunyavirus particle production in Golgi compartments. PLoS Pathog. 2018, 14, e1007047. [Google Scholar] [CrossRef]
- Douam, F.; Hrebikova, G.; Albrecht, Y.E.S.; Sellau, J.; Sharon, Y.; Ding, Q.; Ploss, A. Single-cell tracking of flavivirus RNA uncovers species-specific interactions with the immune system dictating disease outcome. Nat. Commun. 2017, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, S.L.; Tesh, R.B.; Azar, S.R.; Muruato, A.E.; Hanley, K.A.; Auguste, A.J.; Langsjoen, R.M.; Paessler, S.; Vasilakis, N.; Weaver, S.C. Characterization of a Novel Murine Model to Study Zika Virus. Am. J. Trop. Med. Hyg. 2016, 94, 1362–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Correspondence, M.S.D.; Diamond, M.S. A Mouse Model of Zika Virus Pathogenesis Cell Host & Microbe Resource A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe 2016, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Livonesi, M.C.; de Sousa, R.L.M.; Badra, S.J.; Figueiredo, L.T.M. In vitro and in vivo studies of the Interferon-alpha action on distinct Orthobunyavirus. Antivir. Res. 2007, 75, 121–128. [Google Scholar] [CrossRef]
- Montano, M. Model systems. In Translational Biology in Medicine; Elsevier: Amsterdam, The Netherland, 2014; pp. 9–33. ISBN 9781908818652. [Google Scholar] [CrossRef]
- Geddes, V.E.V.; de Oliveira, A.S.; Tanuri, A.; Arruda, E.; Ribeiro-alves, M.; Aguiar, R.S. MicroRNA and cellular targets profiling reveal miR-217 and miR-576-3p as proviral factors during Oropouche infection. PLoS Neglated Trop. Dis. 2018, 12, 1–25. [Google Scholar] [CrossRef]
- Proenca-modena, J.L.; Hyde, J.L.; Sesti-costa, R.; Lucas, T.; Pinto, A.K.; Richner, J.M.; Gorman, M.J.; Lazear, H.M.; Diamond, S. Interferon-Regulatory Factor 5-Dependent Signaling Restricts Orthobunyavirus Dissemination to the Central Nervous System. J. Virol. 2016, 90, 189–205. [Google Scholar] [CrossRef] [Green Version]





| Target | Sequence | Reference |
|---|---|---|
| RIG-1 | F: 5’-TGTGCTCCTACAGGTTGTGGA-3′ R: 5’-CACTGGGATCTGATTCGCAAAA-3′ | [22] |
| MDA5 | F: 5’-CCAAAGCTGAAGAACACAT-3′ R: 5’-ATCTTCTCTGGTTGCATCT-3′ | [23] |
| TRIF | F: 5′-GGCCCATCACTTCCTAGCG-3′ R: 5′-GAGAGATCCTGGCCTCAGTTT-3′ | [24] |
| MAVS | F: 5’-GTCACTTCCTGCTGAGA-3′ R: 5’-TGCTCTGAATTCTCTCCT-3′ | [25] |
| IFNB1 | F: 5’-GCTTGGATTCCTACAAAGAAGCA-3′ R: 5’-ATAGATGGTCAATGCGGCGTC-3’ | [26] |
| IFNA (total) | F: 5′-TCCATGAGVTGATBCAGCAGA-3′ R: 5′-ATTTCTGCTCTGACAACCTCCC-3′ | [27] |
| IFNL1 | F: 5′-CGCCTTGGAAGAGTCACTCA-3′ R: 5′-GAAGCCTCAGGTCCCAATTC-3′ | [28] |
| IFIT1 | F: 5’-GCGCTGGGTATGCGATCTC-3′ R: 5’-CAGCCTGCCTTAGGGGAAG-3′ | [22] |
| IFITM3 | F: 5’-ATGTCGTCTGGTCCCTGTTC-3′ R: 5’-GTCATGAGGATGCCCAGAAT-3′ | [29] |
| MYD88 | F: 5′-ATGGTGGTGGTTGTCTCTGATG-3′ R: 5′-GCATCCTGAGGTTCATCCTGTC-3′ | [30] |
| GAPDH | F: 5′- CCCATGTTCGTCATGGGTGT -3’ R: 5′- TGGTCATGAGTCCTTCCACGATA -3’ | [31] |
| OROV | F: 5′-TACCCA GATGCGATCACCAA-3′ P: 5′-/FAM/ TGCCTTTGGCTGAGGTAAAGGGCTG/36-TAMSp/-3′ R: 5′-TTGCGTCACCATCATTCCAA-3′ | [18] |
| IFNG | F: 5′-TCGGTAACTGACTTGAATGTCCA-3′ R: 5′-TCGCTTCCCTGTTTTAGCTGC-3′ | [32] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro Amorim, M.; Cornejo Pontelli, M.; Fabiano de Souza, G.; Primon Muraro, S.; Toledo-Teixeira, D.A.; Forato, J.; Bispo-dos-Santos, K.; Barbosa, N.S.; Cavalheiro Martini, M.; Lorencini Parise, P.; et al. Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow™ and qRT-PCR Assays. Viruses 2020, 12, 785. https://0-doi-org.brum.beds.ac.uk/10.3390/v12070785
Ribeiro Amorim M, Cornejo Pontelli M, Fabiano de Souza G, Primon Muraro S, Toledo-Teixeira DA, Forato J, Bispo-dos-Santos K, Barbosa NS, Cavalheiro Martini M, Lorencini Parise P, et al. Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow™ and qRT-PCR Assays. Viruses. 2020; 12(7):785. https://0-doi-org.brum.beds.ac.uk/10.3390/v12070785
Chicago/Turabian StyleRibeiro Amorim, Mariene, Marjorie Cornejo Pontelli, Gabriela Fabiano de Souza, Stéfanie Primon Muraro, Daniel A. Toledo-Teixeira, Julia Forato, Karina Bispo-dos-Santos, Natália S. Barbosa, Matheus Cavalheiro Martini, Pierina Lorencini Parise, and et al. 2020. "Oropouche Virus Infects, Persists and Induces IFN Response in Human Peripheral Blood Mononuclear Cells as Identified by RNA PrimeFlow™ and qRT-PCR Assays" Viruses 12, no. 7: 785. https://0-doi-org.brum.beds.ac.uk/10.3390/v12070785