Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core
Abstract
:1. Introduction
2. Methods
2.1. Cloning the Minus 38 Capsid Plasmid
2.2. Ross River Virus (RRV) Capsid Protein Expression and Purification
2.3. Core-Like Particle (CLP) Assembly Assay
2.4. Dynamic Light Scattering (DLS)
2.5. Transmission Electron Microscopy (TEM)
2.6. Agarose gel-Shift Assay
2.7. Differential Scanning Fluorimetry (DSF)
2.8. Purification of Core-Like Particles
2.9. Quantification of Protein by Densitometry
2.10. Quantification of Nucleic Acid by Fluorescence
3. Results
3.1. A Negatively-Charged CP Mutant Eliminates the Need for DNA During CLP Assembly
3.2. WT + M38 CLPs Have Slight Differences in Stability When Compared to WT + DNA CLPs
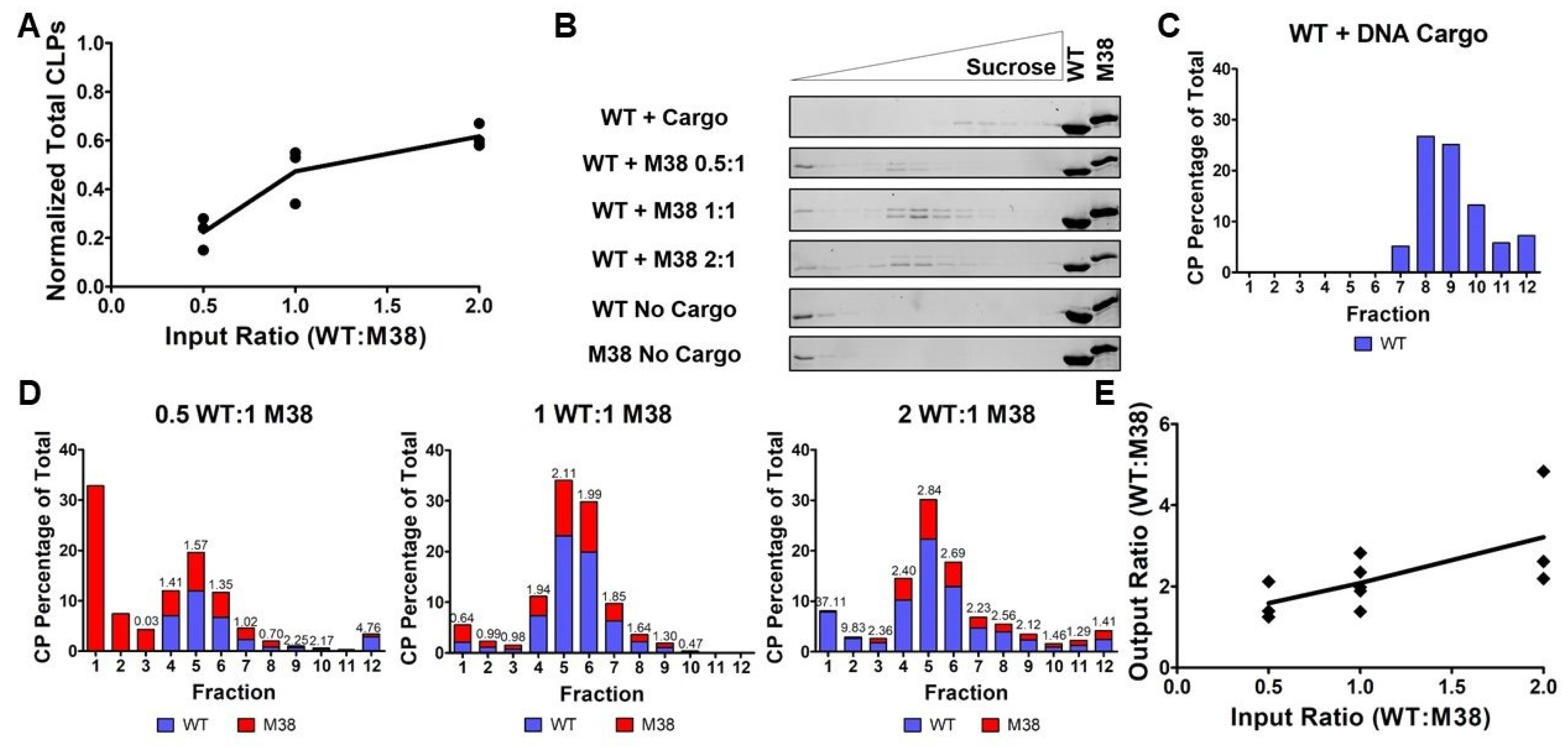
3.3. The Ratio of WT CP and M38 CP Incorporated into CLPs is Determined by input Ratios
3.4. M38 CP Competes with DNA Cargo to Assemble with WT CP into CLPs
3.5. Order of Addition Affects the Composition but not the Quantity of CLPs Assembled
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CP | capsid protein |
| M38 | Minus 38 |
| WT | wild-type |
| CLP | core-like particle |
| DLS | dynamic light scattering |
| TEM | transmission electron microscopy |
| RRV | Ross River Virus |
| DSF | differential scanning fluorimetry |
References
- Chen, R.; Mukhopadhyay, S.; Merits, A.; Bolling, B.; Nasar, F.; Coffey, L.L.; Powers, A.; Weaver, S.C.; ICTV Report Consortium. Ictv virus taxonomy profile: Togaviridae. J. Gen. Virol. 2018, 99, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.H.; Kuhn, R.J.; Olson, N.H.; Rossmann, M.G.; Choi, H.K.; Smith, T.J.; Baker, T.S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 1995, 80, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Jose, J.; Snyder, J.E.; Kuhn, R.J. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009, 4, 837–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Hryc, C.F.; Cong, Y.; Liu, X.; Jakana, J.; Gorchakov, R.; Baker, M.L.; Weaver, S.C.; Chiu, W. 4.4 a cryo-em structure of an enveloped alphavirus venezuelan equine encephalitis virus. EMBO J. 2011, 30, 3854–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Jose, J.; Chipman, P.; Zhang, W.; Kuhn, R.J.; Baker, T.S. Molecular links between the e2 envelope glycoprotein and nucleocapsid core in sindbis virus. J. Mol. Biol. 2011, 414, 442–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostyuchenko, V.A.; Jakana, J.; Liu, X.; Haddow, A.D.; Aung, M.; Weaver, S.C.; Chiu, W.; Lok, S.M. The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 2011, 85, 9327–9333. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, S.; Zhang, W.; Gabler, S.; Chipman, P.R.; Strauss, E.G.; Strauss, J.H.; Baker, T.S.; Kuhn, R.J.; Rossmann, M.G. Mapping the structure and function of the e1 and e2 glycoproteins in alphaviruses. Structure 2006, 14, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Xiang, Y.; Akahata, W.; Holdaway, H.; Pal, P.; Zhang, X.; Diamond, M.S.; Nabel, G.J.; Rossmann, M.G. Structural analyses at pseudo atomic resolution of chikungunya virus and antibodies show mechanisms of neutralization. Elife 2013, 2, e00435. [Google Scholar] [CrossRef]
- Mendes, A.; Kuhn, R.J. Alphavirus nucleocapsid packaging and assembly. Viruses 2018, 10, 138. [Google Scholar] [CrossRef] [Green Version]
- Lulla, V.; Kim, D.Y.; Frolova, E.I.; Frolov, I. The amino-terminal domain of alphavirus capsid protein is dispensable for viral particle assembly but regulates rna encapsidation through cooperative functions of its subdomains. J. Virol. 2013, 87, 12003–12019. [Google Scholar] [CrossRef] [Green Version]
- Forsell, K.; Suomalainen, M.; Garoff, H. Structure-function relation of the nh2-terminal domain of the semliki forest virus capsid protein. J. Virol. 1995, 69, 1556–1563. [Google Scholar] [CrossRef] [Green Version]
- Forsell, K.; Griffiths, G.; Garoff, H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996, 15, 6495–6505. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Przybyla, L.; Edwards, T.J.; Perera, R.; Burgner, J.W., 2nd; Kuhn, R.J. Interactions of the cytoplasmic domain of sindbis virus e2 with nucleocapsid cores promote alphavirus budding. J. Virol. 2012, 86, 2585–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Owen, K.E.; Choi, H.K.; Lee, H.; Lu, G.; Wengler, G.; Brown, D.T.; Rossmann, M.G.; Kuhn, R.J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 1996, 4, 531–541. [Google Scholar] [CrossRef] [Green Version]
- Skoging, U.; Vihinen, M.; Nilsson, L.; Liljestrom, P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 1996, 4, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, T.A.; Tellinghuisen, T.L.; Kuhn, R.J.; Post, C.B. Association of sindbis virus capsid protein with phospholipid membranes and the e2 glycoprotein: Implications for alphavirus assembly. Biochemistry 2005, 44, 2800–2810. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.M.; Perera, R.; Kuhn, R.J. Alphavirus capsid protein helix i controls a checkpoint in nucleocapsid core assembly. J. Virol. 2006, 80, 8848–8855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, R.; Navaratnarajah, C.; Kuhn, R.J. A heterologous coiled coil can substitute for helix i of the sindbis virus capsid protein. J. Virol. 2003, 77, 8345–8353. [Google Scholar] [CrossRef] [Green Version]
- Rayaprolu, V.; Moore, A.; Wang, J.C.; Goh, B.C.; Perilla, J.; Zlotnick, A.; Mukhopadhyay, S. Length of encapsidated cargo impacts stability and structure of in vitro assembled alphavirus core-like particles. J. Phys. Condens Matter 2017, 29, 484003. [Google Scholar] [CrossRef]
- Tellinghuisen, T.L.; Hamburger, A.E.; Fisher, B.R.; Ostendorp, R.; Kuhn, R.J. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in escherichia coli. J. Virol. 1999, 73, 5309–5319. [Google Scholar] [CrossRef] [Green Version]
- Warrier, R.; Linger, B.R.; Golden, B.L.; Kuhn, R.J. Role of sindbis virus capsid protein region ii in nucleocapsid core assembly and encapsidation of genomic rna. J. Virol. 2008, 82, 4461–4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linger, B.R.; Kunovska, L.; Kuhn, R.J.; Golden, B.L. Sindbis virus nucleocapsid assembly: Rna folding promotes capsid protein dimerization. RNA 2004, 10, 128–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tellinghuisen, T.L.; Perera, R.; Kuhn, R.J. In vitro assembly of sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 2001, 75, 2810–2817. [Google Scholar] [CrossRef] [Green Version]
- Perera, R.; Owen, K.E.; Tellinghuisen, T.L.; Gorbalenya, A.E.; Kuhn, R.J. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 2001, 75, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Mukhopadhyay, S. Generating enveloped virus-like particles with in vitro assembled cores. Virology 2011, 413, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Tsvetkova, I.B.; Khuong, Y.L.; Moore, A.W.; Arnold, R.J.; Goicochea, N.L.; Dragnea, B.; Mukhopadhyay, S. The packaging of different cargo into enveloped viral nanoparticles. Mol. Pharm. 2013, 10, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Chen, C.; Rayaprolu, V.; Mukhopadhyay, S.; Zlotnick, A. Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano 2015, 9, 8898–8906. [Google Scholar] [CrossRef] [Green Version]
- Wengler, G.; Wengler, G.; Boege, U.; Wahn, K. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological conditions in vitro. Virology 1984, 132, 401–412. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Chipman, P.R.; Hong, E.M.; Kuhn, R.J.; Rossmann, M.G. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 2002, 76, 11128–11132. [Google Scholar] [CrossRef] [Green Version]
- Snyder, J.E.; Azizgolshani, O.; Wu, B.; He, Y.; Lee, A.C.; Jose, J.; Suter, D.M.; Knobler, C.M.; Gelbart, W.M.; Kuhn, R.J. Rescue of infectious particles from preassembled alphavirus nucleocapsid cores. J. Virol. 2011, 85, 5773–5781. [Google Scholar] [CrossRef] [Green Version]
- Goicochea, N.L.; De, M.; Rotello, V.M.; Mukhopadhyay, S.; Dragnea, B. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 2007, 7, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, P.; Hirth, L. Aggregation states of brome mosaic virus protein. Virology 1974, 61, 160–167. [Google Scholar] [CrossRef]
- Lavelle, L.; Gingery, M.; Phillips, M.; Gelbart, W.M.; Knobler, C.M.; Cadena-Nava, R.D.; Vega-Acosta, J.R.; Pinedo-Torres, L.A.; Ruiz-Garcia, J. Phase diagram of self-assembled viral capsid protein polymorphs. J. Phys. Chem. B 2009, 113, 3813–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atabekov, J.; Nikitin, N.; Arkhipenko, M.; Chirkov, S.; Karpova, O. Thermal transition of native tobacco mosaic virus and rna-free viral proteins into spherical nanoparticles. J. Gen. Virol. 2011, 92, 453–456. [Google Scholar] [CrossRef]
- Bruckman, M.A.; Hern, S.; Jiang, K.; Flask, C.A.; Yu, X.; Steinmetz, N.F. Tobacco mosaic virus rods and spheres as supramolecular high-relaxivity mri contrast agents. J. Mater. Chem. B 2013, 1, 1482–1490. [Google Scholar] [CrossRef] [Green Version]
- Bruckman, M.A.; VanMeter, A.; Steinmetz, N.F. Nanomanufacturing of tobacco mosaic virus-based spherical biomaterials using a continuous flow method. ACS Biomater. Sci. Eng. 2015, 1, 13–18. [Google Scholar] [CrossRef] [Green Version]
- van Eldijk, M.B.; Wang, J.C.; Minten, I.J.; Li, C.; Zlotnick, A.; Nolte, R.J.; Cornelissen, J.J.; van Hest, J.C. Designing two self-assembly mechanisms into one viral capsid protein. J. Am. Chem. Soc. 2012, 134, 18506–18509. [Google Scholar] [CrossRef] [Green Version]
- Ceres, P.; Zlotnick, A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis b virus capsids. Biochemistry 2002, 41, 11525–11531. [Google Scholar] [CrossRef]
- Johnson, M.C.; Scobie, H.M.; Ma, Y.M.; Vogt, V.M. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002, 76, 11177–11185. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. Expasy: Sib bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Zlotnick, A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 2003, 278, 18249–18255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, K.N.; Suhanovsky, M.M.; Teschke, C.M. Phage p22 procapsids equilibrate with free coat protein subunits. J. Mol. Biol. 2007, 365, 513–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liepold, L.; Anderson, S.; Willits, D.; Oltrogge, L.; Frank, J.A.; Douglas, T.; Young, M. Viral capsids as mri contrast agents. Magn. Reson. Med. 2007, 58, 871–879. [Google Scholar] [CrossRef]
- Usselman, R.J.; Qazi, S.; Aggarwal, P.; Eaton, S.S.; Eaton, G.R.; Russek, S.; Douglas, T. Gadolinium-loaded viral capsids as magnetic resonance imaging contrast agents. Appl. Magn. Reson. 2015, 46, 349–355. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Uchida, M.; O’Neil, A.; Li, R.; Prevelige, P.E.; Douglas, T. Implementation of p22 viral capsids as nanoplatforms. Biomacromolecules 2010, 11, 2804–2809. [Google Scholar] [CrossRef]
- Sharma, J.; Uchida, M.; Miettinen, H.M.; Douglas, T. Modular interior loading and exterior decoration of a virus-like particle. Nanoscale 2017, 9, 10420–10430. [Google Scholar] [CrossRef]
- Qazi, S.; Miettinen, H.M.; Wilkinson, R.A.; McCoy, K.; Douglas, T.; Wiedenheft, B. Programmed self-assembly of an active p22-cas9 nanocarrier system. Mol. Pharm. 2016, 13, 1191–1196. [Google Scholar] [CrossRef]
- O’Neil, A.; Prevelige, P.E.; Basu, G.; Douglas, T. Coconfinement of fluorescent proteins: Spatially enforced communication of gfp and mcherry encapsulated within the p22 capsid. Biomacromolecules 2012, 13, 3902–3907. [Google Scholar] [CrossRef]
- Patterson, D.P.; Prevelige, P.E.; Douglas, T. Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage p22. ACS Nano 2012, 6, 5000–5009. [Google Scholar] [CrossRef]
- Patterson, D.P.; Rynda-Apple, A.; Harmsen, A.L.; Harmsen, A.G.; Douglas, T. Biomimetic antigenic nanoparticles elicit controlled protective immune response to influenza. ACS Nano 2013, 7, 3036–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, B.; Morabito, K.M.; Ruckwardt, T.J.; Patterson, D.P.; Avera, J.; Miettinen, H.M.; Graham, B.S.; Douglas, T. Viruslike particles encapsidating respiratory syncytial virus m and m2 proteins induce robust t cell responses. ACS Biomater. Sci. Eng. 2016, 2, 2324–2332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundstrom, K. Self-replicating RNA viruses for RNA therapeutics. Molecules 2018, 23, 3310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundstrom, K. Rna viruses as tools in gene therapy and vaccine development. Genes 2019, 10, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Sample | Ratio | % Sucrose in Peak CLP Fraction * | Normalized Relative CLPs ** |
|---|---|---|---|
| WT + DNA Cargo | 1:1 | 18.68–20.00 | 0.59 |
| WT + M38 | 0.5:1 | 14.59 | 0.26 |
| 1:1 | 14.40 | 0.56 | |
| 2:1 | 14.63 | 0.73 | |
| WT + (M38 + DNA Cargo) | 0.5:1:1 | 17.62 | 0.17 |
| 1:1:1 | 18.93 | 0.40 | |
| 2:1:1 | 18.65 | 1.00 | |
| (WT + DNA Cargo) + M38 | 1:1:1 | 18.94 | 0.42 |
| (WT + M38) + DNA Cargo | 1:1:1 | 17.27 | 0.43 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Button, J.M.; Mukhopadhyay, S. Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core. Viruses 2020, 12, 846. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080846
Button JM, Mukhopadhyay S. Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core. Viruses. 2020; 12(8):846. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080846
Chicago/Turabian StyleButton, Julie M., and Suchetana Mukhopadhyay. 2020. "Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core" Viruses 12, no. 8: 846. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080846