Nationwide Screening for Bee Viruses and Parasites in Belgian Honey Bees
Abstract
:1. Introduction
2. Materials and Methods
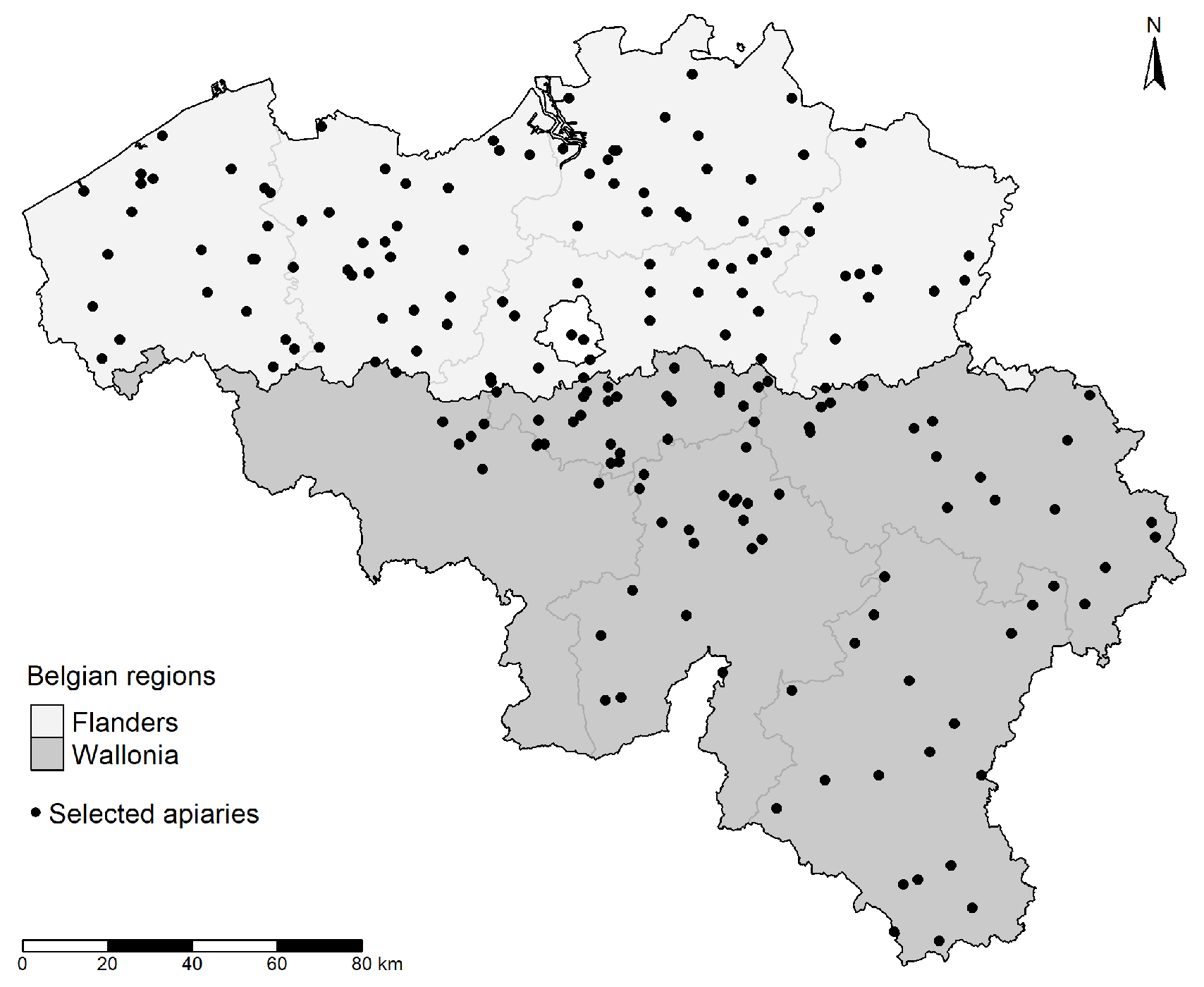
2.1. Honey Bee Sampling
2.2. Virus Detection
2.3. Varroa Mite Counts
2.4. Nosema Spore Counts and Species Identification
2.5. Statistical Analysis
3. Results
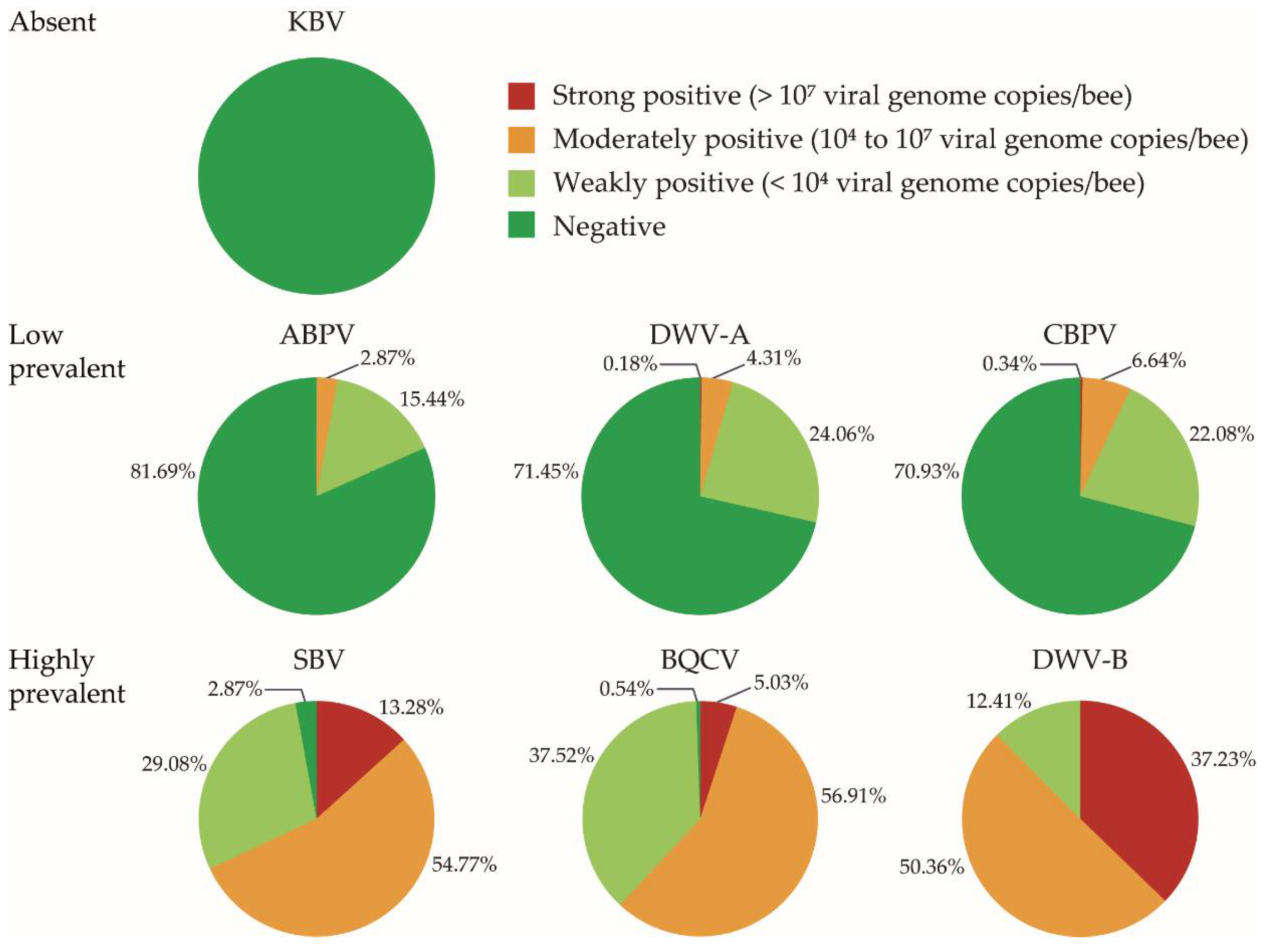
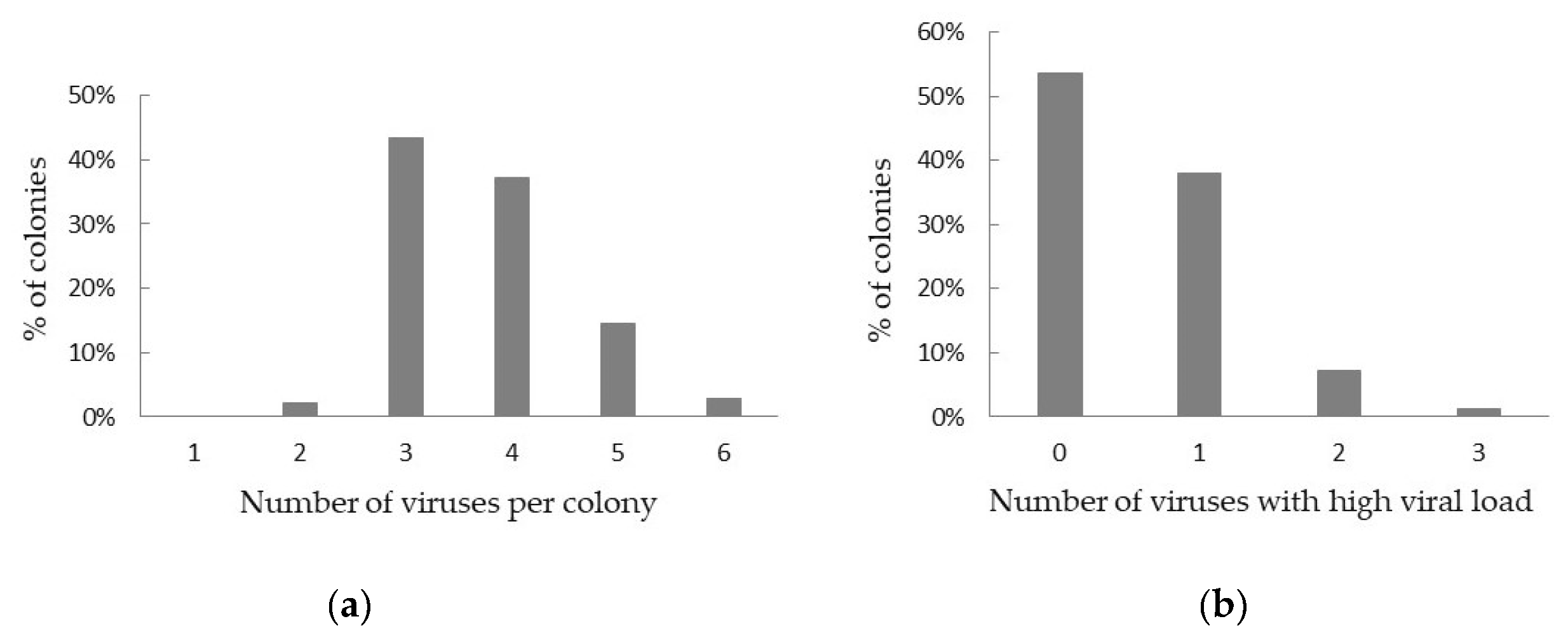
3.1. Virus Prevalence and Viral Load
3.2. Varroa Prevalence and Abundance
3.3. Correlation between Virus Load and Varroa Infestation
3.4. Nosema Prevalence, Abundance and Species Identification
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jacques, A.; Laurent, M.; Ribière-Chabert, M.; Saussac, M.; Bougeard, S.; Budge, G.E.; Hendrikx, P.; Chauzat, M.P. A pan-European epidemiological study reveals honey bee colony survival depends on beekeeper education and disease control. PLoS ONE 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genersch, E.; Von Der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; et al. The German bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef] [Green Version]
- Porrini, C.; Mutinelli, F.; Bortolotti, L.; Granato, A.; Laurenson, L.; Roberts, K.; Gallina, A.; Silvester, N.; Medrzycki, P.; Renzi, T.; et al. The status of honey bee health in Italy: Results from the nationwide bee monitoring network. PLoS ONE 2016, 11, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Charrière, J.D.; Neumann, P. Surveys to estimate winter losses in Switzerland. J. Apic. Res. 2010, 49, 132–133. [Google Scholar] [CrossRef]
- Spleen, A.M.; Lengerich, E.J.; Rennich, K.; Caron, D.; Rose, R.; Pettis, J.S.; Henson, M.; Wilkes, J.T.; Wilson, M.; Stitzinger, J.; et al. A national survey of managed honey bee 2011-12 winter colony losses in the United States: Results from the Bee Informed Partnership. J. Apic. Res. 2013, 52, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noël, L.M.L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009. [Google Scholar] [CrossRef] [Green Version]
- Berthoud, H.; Imdorf, A.; Haueter, M.; Radloff, S.; Neumann, P. Virus infections and winter losses of honey bee colonies (Apis mellifera). J. Apic. Res. 2010, 49, 60–65. [Google Scholar] [CrossRef]
- Iqbal, J.; Mueller, U. Virus infection causes specific learning deficits in honeybee foragers. Proc. R. Soc. B Biol. Sci. 2007, 274, 1517–1521. [Google Scholar] [CrossRef] [Green Version]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumanna, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 2012, 78, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Beaurepaire, A.; Piot, N.; Doublet, V.; Antunez, K.; Campbell, E.; Chantawannakul, P.; Chejanovsky, N.; Gajda, A.; Heerman, M.; Panziera, D.; et al. Diversity and Global Distribution of Viruses of the Western Honey Bee, Apis mellifera. Insects 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.-P.; Gauthier, L.; Genersch, E.; de Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Evans, J.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Amiri, E.; Meixner, M.D.; Kryger, P. Deformed wing virus can be transmitted during natural mating in honey bees and infect the queens. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pettis, J.S.; Evans, J.D.; Kramer, M.; Feldlaufer, M.F. Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 2004, 35, 441–448. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Yang, X.; Cox-Foster, D.; Cui, L. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 2005, 342, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Gisder, S.; Aumeier, P.; Genersch, E. Deformed wing virus: Replication and viral load in mites (Varroa destructor). J. Gen. Virol. 2009, 90, 463–467. [Google Scholar] [CrossRef]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef]
- Ball, B.V.; Allen, M.F. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 1988, 113, 237–244. [Google Scholar] [CrossRef]
- Bowen-Walker, P.L.; Martin, S.J.; Gunn, A. The Transmission of Deformed Wing Virus between Honeybees (Apis mellifera L.) by the Ectoparasitic Mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 1999, 73, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Genersch, E. Honey bee pathology: Current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 2010, 87, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Naggar, Y.; Paxton, R.J. Mode of Transmission Determines the Virulence of Black Queen Cell Virus in Adult Honey Bees, Posing a Future Threat to Bees and Apiculture. Viruses 2020, 12, 535. [Google Scholar] [CrossRef] [PubMed]
- Chapter 3.2.4. Nosemosis of honey bees. In OIE Terrestrial Manual; 2018; p. 1833. Available online: https://www.oie.int/standard-setting/terrestrial-manual/access-online/ (accessed on 14 August 2020).
- Toplak, I.; Jamnikar Ciglenečki, U.; Aronstein, K.; Gregorc, A. Chronic bee paralysis virus and Nosema ceranae experimental co-infection of winter honey bee workers (Apis mellifera L.). Viruses 2013, 5, 2282–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doublet, V.; Natsopoulou, M.E.; Zschiesche, L.; Paxton, R.J. Within-host competition among the honey bees pathogens Nosema ceranae and Deformed wing virus is asymmetric and to the disadvantage of the virus. J. Invertebr. Pathol. 2015, 124, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Doublet, V.; Labarussias, M.; de Miranda, J.R.; Moritz, R.F.A.; Paxton, R.J. Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 2015, 17, 969–983. [Google Scholar] [CrossRef]
- Nguyen, B.K.; Ribière, M.; VanEngelsdorp, D.; Snoeck, C.; Saegerman, C.; Kalkstein, A.L.; Schurr, F.; Brostaux, Y.; Faucon, J.-P.; Haubruge, E. Effects of honey bee virus prevalence, Varroa destructor load and queen condition on honey bee colony survival over the winter in Belgium. J. Apic. Res. 2011, 50, 195–202. [Google Scholar] [CrossRef]
- De Smet, L.; Ravoet, J.; de Miranda, J.R.; Wenseleers, T.; Mueller, M.Y.; Moritz, R.F.A.; de Graaf, D.C. BeeDoctor, a Versatile MLPA-Based Diagnostic Tool for Screening Bee Viruses. PLoS ONE 2012, 7, e47953. [Google Scholar] [CrossRef] [Green Version]
- Forsgren, E.; Locke, B.; Semberg, E.; Laugen, A.T.; de Miranda, J.R. Sample preservation, transport and processing strategies for honeybee RNA extraction: Influence on RNA yield, quality, target quantification and data normalization. J. Virol. Methods 2017, 246, 81–89. [Google Scholar] [CrossRef]
- Ciglenečki, U.J.; Toplak, I. Development of a real-time RT-PCR assay with TaqMan probe for specific detection of acute bee paralysis virus. J. Virol. Methods 2012, 184, 63–68. [Google Scholar] [CrossRef]
- Chantawannakul, P.; Ward, L.; Boonham, N.; Brown, M. A scientific note on the detection of honeybee viruses using real-time PCR (TaqMan) in Varroa mites collected from a Thai honeybee (Apis mellifera) apiary. J. Invertebr. Pathol. 2006, 91, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.; Ribière, M.; Celle, O.; Lallemand, P.; Schurr, F.; Olivier, V.; Iscache, A.L.; Faucon, J.P. Evaluation of a real-time two-step RT-PCR assay for quantitation of Chronic bee paralysis virus (CBPV) genome in experimentally-infected bee tissues and in life stages of a symptomatic colony. J. Virol. Methods 2007, 141, 7–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.P.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of quantitative real-time RT-PCR assays for the detection of six honeybee viruses. J. Virol. Methods 2019, 270, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.; Waite, R.; Boonham, N.; Fisher, T.; Pescod, K.; Thompson, H.; Chantawannakul, P.; Brown, M. First detection of Kashmir bee virus in the UK using real-time PCR. Apidologie 2007, 38, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.P.; Thiéry, R.; Ribière, M. Development and validation of a real-time two-step RT-qPCR TaqMan® assay for quantitation of Sacbrood virus (SBV) and its application to a field survey of symptomatic honey bee colonies. J. Virol. Methods 2014, 197, 7–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.P.; Higgins, J.A.; Feldlaufer, M.F. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 2005, 71, 436–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D.; vanEngelsdorp, D. Multiyear survey targeting disease incidence in US honey bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef] [Green Version]
- Ravoet, J.; De Smet, L.; Wenseleers, T.; de Graaf, D.C. Vertical transmission of honey bee viruses in a Belgian queen breeding program. BMC Vet. Res. 2015, 11. [Google Scholar] [CrossRef] [Green Version]
- Schittny, D.; Yañez, O.; Neumann, P. Honey Bee Virus Transmission via Hive Products. Vet. Sci. 2020, 7, 96. [Google Scholar] [CrossRef]
- Mutinelli, F. The spread of pathogens through trade in honey bees and their products (including queen bees and semen): Overview and recent developments. Rev. Sci. Tech. l’OIE 2011, 30, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and Sacbrood virus) with the honeybee host and the parasitic Varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Ravoet, J.; De Smet, L.; Meeus, I.; Smagghe, G.; Wenseleers, T.; de Graaf, D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E.; Yue, C.; Fries, I.; De Miranda, J.R. Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J. Invertebr. Pathol. 2006, 91, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Bailes, E.J.; Deutsch, K.R.; Bagi, J.; Rondissone, L.; Brown, M.J.F.; Owen, T. First detection of honey bee viruses in hoverfly (syrphid) pollinators. Biol. Lett. 2016, 14, 20180001. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cox-Foster, D.L. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 2005, 102, 7470–7475. [Google Scholar] [CrossRef] [Green Version]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [Green Version]
- Biesmeijer, K. Report Honeybee Surveillance Program the Netherlands. Naturalis 2016. [Google Scholar]
- Amiri, E.; Meixner, M.; Nielsen, S.L.; Kryger, P. Four categories of viral infection describe the health status of honey bee colonies. PLoS ONE 2015, 10, 10–12. [Google Scholar] [CrossRef] [Green Version]
- Benaets, K.; Van Geystelen, A.; Cardoen, D.; De Smet, L.; de Graaf, D.C.; Schoofs, L.; Larmuseau, M.H.D.; Brettell, L.E.; Martin, S.J.; Wenseleers, T. Covert deformed wing virus infections have long-term deleterious effects on honeybee foraging and survival. Proc. R. Soc. B Biol. Sci. 2017, 284. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018. [Google Scholar] [CrossRef]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; de Graaf, D.C. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milbrath, M.O.; van Tran, T.; Huang, W.F.; Solter, L.F.; Tarpy, D.R.; Lawrence, F.; Huang, Z.Y. Comparative virulence and competition between Nosema apis and Nosema ceranae in honey bees (Apis mellifera). J. Invertebr. Pathol. 2015, 125, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Bryden, J.; Paxton, R.J. Interspecific competition in honeybee intracellular gut parasites is asymmetric and favours the spread of an emerging infectious disease. Proc. R. Soc. B Biol. Sci. 2015, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Virus | Sequences Primers and Probe (5′–3′) | Reference | Final Primer—Probe Concentration |
|---|---|---|---|
| ABPV | Forward CATATTGGCGAGCCACTATG | [31] | 800 nM—100 nM |
| Reverse CTACCAGGTTCAAAGAAAATTTC | |||
| Probe ATAGTTAAAACAGCTTTTCACACTGG | |||
| BQCV | Forward GGTGCGGGAGATGATATGGA | [32] | 320 nM—200 nM |
| Reverse GCCGTCTGAGATGCATGAATAC | |||
| Probe TTTCCATCTTTATCGGTACGCCGCC | |||
| CBPV | Forward CGCAAGTACGCCTTGATAAAGAAC | [33] | 320 nM—200 nM |
| Reverse ACTACTAGAAACTCGTCGCTTCG | |||
| Probe TCAAGAACGAGACCACCGCCAAGTTC | |||
| DWV-A | Forward GCGGCTAAGATTGTAAATTG | [34] | 350 nM—100 nM |
| Reverse GTGACTAGCATAACCATGATTA | |||
| Probe CCTTGACCAGTAGACACAGCATC | |||
| DWV-B | Forward GGTCTGAAGCGAAAATAG | [34] | 600 nM—200 nM |
| Reverse CTAGCATATCCATGATTATAAAC | |||
| Probe CCTTGTCCAGTAGATACAGCATCACA | |||
| KBV | Forward ACCAGGAAGTATTCCCATGGTAAG | [35] | 500 nM—200 nM |
| Reverse TGGAGCTATGGTTCCGTTCAG | |||
| Probe CCGCAGATAACTTAGGACCAGATCAATCACA | |||
| SBV | Forward AACGTCCACTACACCGAAATGTC | [36] | 320 nM—200 nM |
| Reverse ACACTGCGCGTCTAACATTCC | |||
| Probe TGATGAGAGTGGACGAAGA | |||
| actin | Forward AGGAATGGAAGCTTGCGGTA | [37] | 500 nM—200 nM |
| Reverse AATTTTCATGGTGGATGGTGC | |||
| Probe ATGCCAACACTGTCCTTTCTGGAGGTA |
| Nosema Species | Sequences Primers and Probe (5′–3′) | Reference | Final Primer—Probe Concentration |
|---|---|---|---|
| Nosema apis | Forward CCATTGCCGGATAAGAGAGT | [38] | 300 nM—150 nM |
| Reverse CCACCAAAAACTCCCAAGAG | |||
| Probe ATAGTGAGGCTCTATCACTCCGCTG | |||
| Nosema ceranae | Forward CGGATAAAAGAGTCCGTTACC | [38] | 300 nM—150 nM |
| Reverse TGAGCAGGGTTCTAGGGAT | |||
| Probe CGTTACCCTTCGGGGAATCTTC |
| Virus | Percentage of Positive Colonies in Flanders | Trend 1 | Percentage of Positive Colonies in Wallonia | Trend 2 |
|---|---|---|---|---|
| ABPV | 8.8% | + | 26.5% | + |
| BQCV | 99.23% | + | 99.65% | + |
| CBPV | 29.23% | + | 30.27% | - |
| DWV-A | 18.07% | + 3 | 37.75% | + 3 |
| DWV-B | 100% | 100% | ||
| KBV | 0 | no data | 0 | no data |
| SBV | 97.69% | + | 96.26% | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matthijs, S.; De Waele, V.; Vandenberge, V.; Verhoeven, B.; Evers, J.; Brunain, M.; Saegerman, C.; De Winter, P.J.J.; Roels, S.; de Graaf, D.C.; et al. Nationwide Screening for Bee Viruses and Parasites in Belgian Honey Bees. Viruses 2020, 12, 890. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080890
Matthijs S, De Waele V, Vandenberge V, Verhoeven B, Evers J, Brunain M, Saegerman C, De Winter PJJ, Roels S, de Graaf DC, et al. Nationwide Screening for Bee Viruses and Parasites in Belgian Honey Bees. Viruses. 2020; 12(8):890. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080890
Chicago/Turabian StyleMatthijs, Severine, Valérie De Waele, Valerie Vandenberge, Bénédicte Verhoeven, Jacqueline Evers, Marleen Brunain, Claude Saegerman, Paul J. J. De Winter, Stefan Roels, Dirk C. de Graaf, and et al. 2020. "Nationwide Screening for Bee Viruses and Parasites in Belgian Honey Bees" Viruses 12, no. 8: 890. https://0-doi-org.brum.beds.ac.uk/10.3390/v12080890





