Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure
Abstract
:1. Introduction
2. Receptor Binding of Influenza HA
3. HA Fusion Machinery and Mechanism
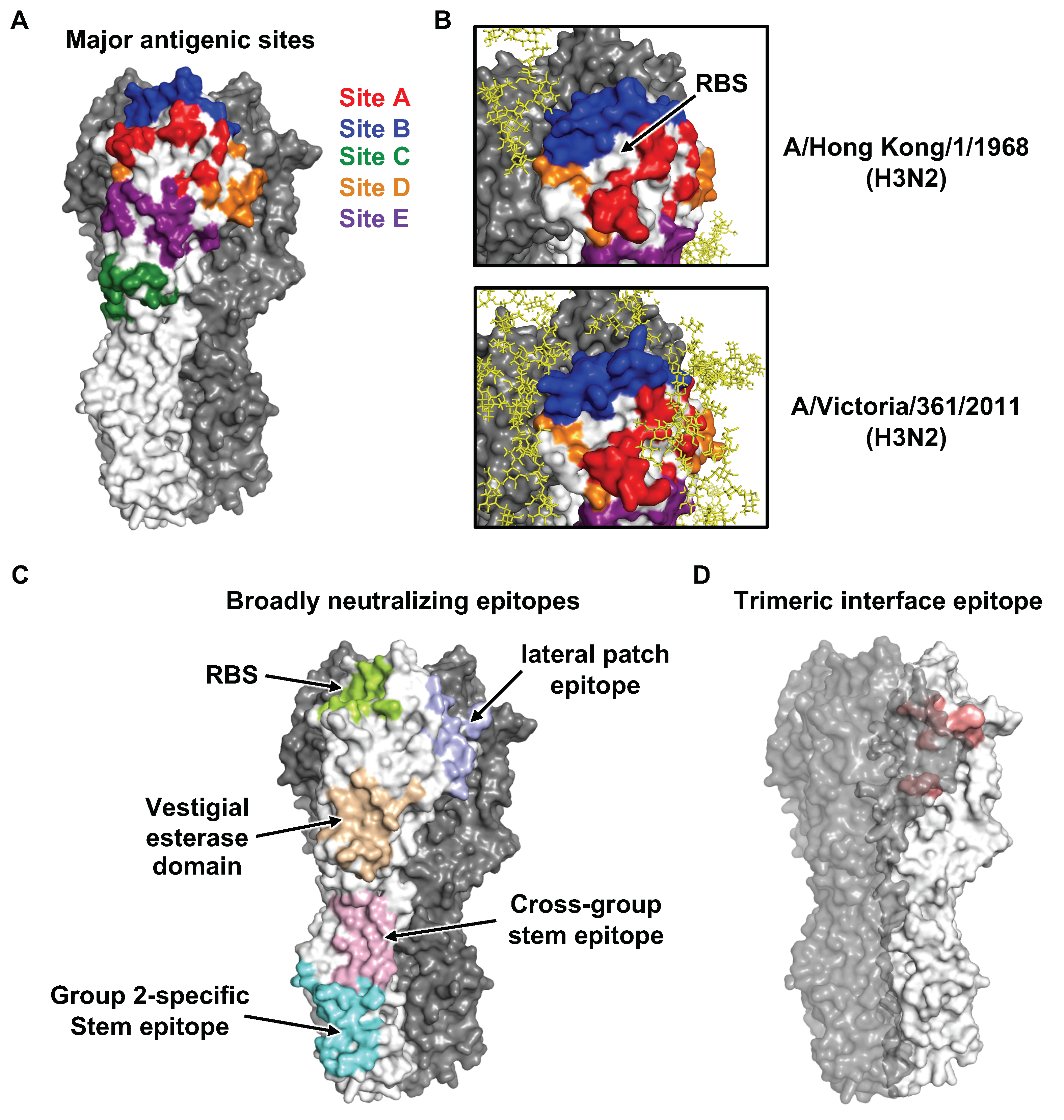
4. Antibodies to Influenza HA
5. HA-Based Therapeutic and Vaccine Design
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.W.; Xu, X.; Li, Y.; Normand, S.; Ueki, R.T.; Kunimoto, G.Y.; Hall, H.; Klimov, A.; Cox, N.J.; Subbarao, K. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000–2001 and 2001–2002 seasons. Virology 2002, 303, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Veit, M. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein Cell 2016, 7, 28–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowdle, W.R. Influenza A virus recycling revisited. Bull. World Health Organ. 1999, 77, 820–828. [Google Scholar]
- CDC. H5N1 in People. Available online: https://www.cdc.gov/flu/avianflu/h5n1-people.htm (accessed on 30 April 2020).
- WHO. Recommended Composition of Influenza Virus Vaccines for Use in the 2019–2020 Northern Hemisphere Influenza Season. Available online: https://www.who.int/influenza/vaccines/virus/recommendations/2019_20_north/en/ (accessed on 29 August 2020).
- Belongia, E.A.; Simpson, M.D.; King, J.P.; Sundaram, M.E.; Kelley, N.S.; Osterholm, M.T.; McLean, H.Q. Variable influenza vaccine effectiveness by subtype: A systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016, 16, 942–951. [Google Scholar] [CrossRef]
- Krammer, F.; Fouchier, R.A.M.; Eichelberger, M.C.; Webby, R.J.; Shaw-Saliba, K.; Wan, H.; Wilson, P.C.; Compans, R.W.; Skountzou, I.; Monto, A.S. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 2018, 9, e02332-17. [Google Scholar] [CrossRef] [Green Version]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Hirst, G.K. The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science 1941, 94, 22–23. [Google Scholar] [CrossRef]
- Hirst, G.K. Adsorption of influenza hemagglutinins and virus by red blood cells. J. Exp. Med. 1942, 76, 195–209. [Google Scholar] [CrossRef] [Green Version]
- Burnet, F.M.; Stone, J.D. The receptor-destroying enzyme of V. cholerae. Aust. J. Exp. Biol. Med. Sci. 1947, 25, 227–233. [Google Scholar] [CrossRef]
- Stone, J.D. Prevention of virus infection with enzyme of V. cholerae, studies with influenza virus in mice. Aust. J. Exp. Biol. Med. Sci. 1948, 26, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Klenk, E.; Faillard, H.; Lempfrid, H. Enzymatic effect of the influenza virus. Hoppe. Seylers Z. Physiol. Chem. 1955, 301, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.S. Crystallographic elucidation of the saccharide binding mode in wheat germ agglutinin and its biological significance. J. Mol. Biol. 1980, 141, 267–291. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C.; Daniels, R.S.; Skehel, J.J.; Wilson, I.A.; Wiley, D.C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 1983, 304, 76–78. [Google Scholar] [CrossRef]
- Weis, W.; Brown, J.H.; Cusack, S.; Paulson, J.C.; Skehel, J.J.; Wiley, D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, X.; Chen, X.; Ma, J. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc. Natl. Acad. Sci. USA 2007, 104, 16874–16879. [Google Scholar] [CrossRef] [Green Version]
- Bradley, K.C.; Galloway, S.E.; Lasanajak, Y.; Song, X.; Heimburg-Molinaro, J.; Yu, H.; Chen, X.; Talekar, G.R.; Smith, D.F.; Cummings, R.D.; et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J. Virol. 2011, 85, 12387–12398. [Google Scholar] [CrossRef] [Green Version]
- Broszeit, F.; Tzarum, N.; Zhu, X.; Nemanichvili, N.; Eggink, D.; Leenders, T.; Li, Z.; Liu, L.; Wolfert, M.A.; Papanikolaou, A.; et al. N-glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep. 2019, 27, 3284–3294. [Google Scholar] [CrossRef] [Green Version]
- Wasik, B.R.; Barnard, K.N.; Parrish, C.R. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 2016, 24, 991–1001. [Google Scholar] [CrossRef]
- Rogers, G.N.; Herrler, G.; Paulson, J.C.; Klenk, H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986, 261, 5947–5951. [Google Scholar]
- Rosenthal, P.B.; Zhang, X.; Formanowski, F.; Fitz, W.; Wong, C.H.; Meier-Ewert, H.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 1998, 396, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified influenza D virus: Basis for its broad cell tropism. PLoS Pathog. 2016, 12, e1005411. [Google Scholar]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Shi, Y.; Wu, Y.; Zhang, W.; Qi, J.; Gao, G.F. Enabling the ‘host jump’: Structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 2014, 12, 822–831. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Gambaryan, A.S.; Teneberg, S.; Piskarev, V.E.; Yamnikova, S.S.; Lvov, D.K.; Robertson, J.S.; Karlsson, K.A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 1997, 233, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Glaser, L.; Stevens, J.; Zamarin, D.; Wilson, I.A.; Garcia-Sastre, A.; Tumpey, T.M.; Basler, C.F.; Taubenberger, J.K.; Palese, P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 2005, 79, 11533–11536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, J.; Blixt, O.; Glaser, L.; Taubenberger, J.K.; Palese, P.; Paulson, J.C.; Wilson, I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 2006, 355, 1143–1155. [Google Scholar] [CrossRef]
- Tumpey, T.M.; Maines, T.R.; Van Hoeven, N.; Glaser, L.; Solorzano, A.; Pappas, C.; Cox, N.J.; Swayne, D.E.; Palese, P.; Katz, J.M.; et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 2007, 315, 655–659. [Google Scholar] [CrossRef]
- Xu, R.; McBride, R.; Nycholat, C.M.; Paulson, J.C.; Wilson, I.A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 2012, 86, 982–990. [Google Scholar] [CrossRef] [Green Version]
- Connor, R.J.; Kawaoka, Y.; Webster, R.G.; Paulson, J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994, 205, 17–23. [Google Scholar] [CrossRef]
- Pappas, C.; Viswanathan, K.; Chandrasekaran, A.; Raman, R.; Katz, J.M.; Sasisekharan, R.; Tumpey, T.M. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS ONE 2010, 5, e11158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; McBride, R.; Paulson, J.C.; Basler, C.F.; Wilson, I.A. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol. 2010, 84, 1715–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, H.; Perez, D.R. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. 2007, 81, 5181–5191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Wang, J.; Bi, Y.; Fan, D.; Liu, H.; Luo, N.; Yang, Z.; Wang, S.; Chen, W.; Wang, J.; et al. Characterization of avian influenza H9N2 viruses isolated from ostriches (Struthio camelus). Sci. Rep. 2018, 8, 2273. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Qi, J.; Xiao, H.; Bi, Y.; Zhang, W.; Xu, Y.; Wang, F.; Shi, Y.; Gao, G.F. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep. 2017, 20, 1201–1214. [Google Scholar] [CrossRef] [Green Version]
- Herfst, S.; Schrauwen, E.J.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 2012, 336, 1534–1541. [Google Scholar] [CrossRef] [Green Version]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Tzarum, N.; Peng, W.; Thompson, A.J.; Ambepitiya Wickramasinghe, I.N.; de la Pena, A.T.T.; van Breemen, M.J.; Bouwman, K.M.; Zhu, X.; McBride, R.; et al. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol. Med. 2017, 9, 1314–1325. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Peng, W.; Grant, O.C.; Thompson, A.J.; Zhu, X.; Bouwman, K.M.; de la Pena, A.T.T.; van Breemen, M.J.; Ambepitiya Wickramasinghe, I.N.; de Haan, C.A.M.; et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog. 2017, 13, e1006390. [Google Scholar] [CrossRef] [Green Version]
- Tzarum, N.; de Vries, R.P.; Peng, W.; Thompson, A.J.; Bouwman, K.M.; McBride, R.; Yu, W.; Zhu, X.; Verheije, M.H.; Paulson, J.C.; et al. The 150-loop restricts the host specificity of human H10N8 influenza virus. Cell Rep. 2017, 19, 235–245. [Google Scholar] [CrossRef]
- Gamblin, S.J.; Haire, L.F.; Russell, R.J.; Stevens, D.J.; Xiao, B.; Ha, Y.; Vasisht, N.; Steinhauer, D.A.; Daniels, R.S.; Elliot, A.; et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 2004, 303, 1838–1842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Viswanathan, K.; Raman, R.; Yu, W.; Sasisekharan, R.; Wilson, I.A. Structural basis for a switch in receptor binding specificity of two H5N1 hemagglutinin mutants. Cell Rep. 2015, 13, 1683–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.; de Vries, R.P.; Grant, O.C.; Thompson, A.J.; McBride, R.; Tsogtbaatar, B.; Lee, P.S.; Razi, N.; Wilson, I.A.; Woods, R.J.; et al. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 2017, 21, 23–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.P.; Xiong, X.; Wharton, S.A.; Martin, S.R.; Coombs, P.J.; Vachieri, S.G.; Christodoulou, E.; Walker, P.A.; Liu, J.; Skehel, J.J.; et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl. Acad. Sci. USA 2012, 109, 21474–21479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.C.; Thompson, A.J.; Xie, J.; Lin, C.W.; Nycholat, C.M.; Zhu, X.; Lerner, R.A.; Paulson, J.C.; Wilson, I.A. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat. Commun. 2018, 9, 1264. [Google Scholar] [CrossRef]
- Wu, N.C.; Otwinowski, J.; Thompson, A.J.; Nycholat, C.M.; Nourmohammad, A.; Wilson, I.A. Major antigenic site B of human influenza H3N2 viruses has an evolving local fitness landscape. Nat. Commun. 2020, 11, 1233. [Google Scholar] [CrossRef]
- Wu, N.C.; Zost, S.J.; Thompson, A.J.; Oyen, D.; Nycholat, C.M.; McBride, R.; Paulson, J.C.; Hensley, S.E.; Wilson, I.A. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog. 2017, 13, e1006682. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Yu, W.; McBride, R.; Li, Y.; Chen, L.M.; Donis, R.O.; Tong, S.; Paulson, J.C.; Wilson, I.A. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. USA 2013, 110, 1458–1463. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [Green Version]
- Karakus, U.; Thamamongood, T.; Ciminski, K.; Ran, W.; Gunther, S.C.; Pohl, M.O.; Eletto, D.; Jeney, C.; Hoffmann, D.; Reiche, S.; et al. MHC class II proteins mediate cross-species entry of bat influenza viruses. Nature 2019, 567, 109–112. [Google Scholar] [CrossRef]
- Giotis, E.S.; Carnell, G.; Young, E.F.; Ghanny, S.; Soteropoulos, P.; Wang, L.F.; Barclay, W.S.; Skinner, M.A.; Temperton, N. Entry of the bat influenza H17N10 virus into mammalian cells is enabled by the MHC class II HLA-DR receptor. Nat. Microbiol. 2019, 4, 2035–2038. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, K.H.; Steinhauer, D.A.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 1998, 95, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Steinhauer, D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 1999, 258, 1–20. [Google Scholar] [CrossRef]
- Skehel, J.J.; Bayley, P.M.; Brown, E.B.; Martin, S.R.; Waterfield, M.D.; White, J.M.; Wilson, I.A.; Wiley, D.C. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. USA 1982, 79, 968–972. [Google Scholar] [CrossRef] [Green Version]
- Ruigrok, R.W.; Aitken, A.; Calder, L.J.; Martin, S.R.; Skehel, J.J.; Wharton, S.A.; Weis, W.; Wiley, D.C. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 1988, 69, 2785–2795. [Google Scholar] [CrossRef]
- Bullough, P.A.; Hughson, F.M.; Skehel, J.J.; Wiley, D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 1994, 371, 37–43. [Google Scholar] [CrossRef]
- Benton, D.J.; Gamblin, S.J.; Rosenthal, P.B.; Skehel, J.J. Structural transitions in influenza haemagglutinin at membrane fusion pH. Nature 2020, 583, 150–153. [Google Scholar] [CrossRef]
- Chen, J.; Skehel, J.J.; Wiley, D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 1999, 96, 8967–8972. [Google Scholar] [CrossRef] [Green Version]
- Bottcher, C.; Ludwig, K.; Herrmann, A.; van Heel, M.; Stark, H. Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 1999, 463, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Calder, L.J.; Rosenthal, P.B. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat. Struct. Mol. Biol. 2016, 23, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wilson, I.A. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 2011, 85, 5172–5182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y. Single-particle cryo-EM-How did it get here and where will it go. Science 2018, 361, 876–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benton, D.J.; Nans, A.; Calder, L.J.; Turner, J.; Neu, U.; Lin, Y.P.; Ketelaars, E.; Kallewaard, N.L.; Corti, D.; Lanzavecchia, A.; et al. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 2018, 115, 10112–10117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhoeyen, M.; Fang, R.; Jou, W.M.; Devos, R.; Huylebroeck, D.; Saman, E.; Fiers, W. Antigenic drift between the haemagglutinin of the Hong Kong influenza strains A/Aichi/2/68 and A/Victoria/3/75. Nature 1980, 286, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Jou, W.M.; Verhoeyen, M.; Devos, R.; Saman, E.; Fang, R.; Huylebroeck, D.; Fiers, W.; Threlfall, G.; Barber, C.; Carey, N.; et al. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell 1980, 19, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Laver, W.G.; Air, G. Structure and variation in influenza virus. In Proceedings of the International Workshop on Structure and Variation in Influenza Virus, Thredbo, Australia, 10–12 December 1979; Elsevier/North-Holland: New York, NY, USA, 1980; p. 395. [Google Scholar]
- Burnet, F.M. Principles of Animal Virology; Academic Press: New York, NY, USA, 1955; p. 486. [Google Scholar]
- Pereira, M.S. Global surveillance of influenza. Br. Med. Bull. 1979, 35, 9–14. [Google Scholar] [CrossRef]
- Stuart-Harris, C. Epidemiology of influenza in man. Br. Med. Bull. 1979, 35, 3–8. [Google Scholar] [CrossRef]
- Fazekas, S.; Groth, S. Antigenic, adaptive and adsorptive variants of the influenza A hemagglutinin. In The Influenza Virus Hemagglutinin; Springer Science and Business Media LLC.: Vienna, Austria, 1978; pp. 25–48. [Google Scholar]
- Laver, W.G.; Air, G.M.; Webster, R.G.; Gerhard, W.; Ward, C.W.; Dopheide, T.A. Antigenic drift in type A influenza virus: Sequence differences in the hemagglutinin of Hong Kong (H3N2) variants selected with monoclonal hybridoma antibodies. Virology 1979, 98, 226–237. [Google Scholar] [CrossRef]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef]
- Skehel, J.J.; Stevens, D.J.; Daniels, R.S.; Douglas, A.R.; Knossow, M.; Wilson, I.A.; Wiley, D.C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 1984, 81, 1779–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef] [PubMed]
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31, 417–427. [Google Scholar] [CrossRef]
- Gerhard, W.; Yewdell, J.; Frankel, M.E.; Webster, R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 1981, 290, 713–717. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, F.; Lu, M.; Tian, X.; Ma, J. Crystal structure of unliganded influenza B virus hemagglutinin. J. Virol. 2008, 82, 3011–3020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Kang, D.S.; Zheng, A.; Liu, S.T.H.; Broecker, F.; Simon, V.; Krammer, F.; Palese, P. Antibody responses toward the major antigenic sites of influenza B virus hemagglutinin in mice, ferrets, and humans. J. Virol. 2019, 93, e01673-18. [Google Scholar] [CrossRef] [Green Version]
- Bizebard, T.; Gigant, B.; Rigolet, P.; Rasmussen, B.; Diat, O.; Bosecke, P.; Wharton, S.A.; Skehel, J.J.; Knossow, M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 1995, 376, 92–94. [Google Scholar] [CrossRef]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Koel, B.F.; Burke, D.F.; Bestebroer, T.M.; van der Vliet, S.; Zondag, G.C.; Vervaet, G.; Skepner, E.; Lewis, N.S.; Spronken, M.I.; Russell, C.A.; et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013, 342, 976–979. [Google Scholar] [CrossRef]
- Li, Y.; Bostick, D.L.; Sullivan, C.B.; Myers, J.L.; Griesemer, S.B.; Stgeorge, K.; Plotkin, J.B.; Hensley, S.E. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J. Virol. 2013, 87, 9904–9910. [Google Scholar] [CrossRef] [Green Version]
- Hensley, S.E.; Das, S.R.; Bailey, A.L.; Schmidt, L.M.; Hickman, H.D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J.R.; et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 2009, 326, 734–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zost, S.J.; Parkhouse, K.; Gumina, M.E.; Kim, K.; Diaz Perez, S.; Wilson, P.C.; Treanor, J.J.; Sant, A.J.; Cobey, S.; Hensley, S.E. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. USA 2017, 114, 12578–12583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.C.; Lv, H.; Thompson, A.J.; Wu, D.C.; Ng, W.W.S.; Kadam, R.U.; Lin, C.W.; Nycholat, C.M.; McBride, R.; Liang, W.; et al. Preventing an antigenically disruptive mutation in egg-based H3N2 seasonal influenza vaccines by mutational incompatibility. Cell Host Microbe 2019, 25, 836–844.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond, D.D.; Stewart, S.M.; Lee, J.; Ferdman, J.; Bajic, G.; Do, K.T.; Ernandes, M.J.; Suphaphiphat, P.; Settembre, E.C.; Dormitzer, P.R.; et al. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat. Med. 2016, 22, 1465–1469. [Google Scholar] [CrossRef]
- Abe, Y.; Takashita, E.; Sugawara, K.; Matsuzaki, Y.; Muraki, Y.; Hongo, S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J. Virol. 2004, 78, 9605–9611. [Google Scholar] [CrossRef] [Green Version]
- Das, S.R.; Puigbo, P.; Hensley, S.E.; Hurt, D.E.; Bennink, J.R.; Yewdell, J.W. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010, 6, e1001211. [Google Scholar] [CrossRef]
- Altman, M.O.; Angel, M.; Kosik, I.; Trovao, N.S.; Zost, S.J.; Gibbs, J.S.; Casalino, L.; Amaro, R.E.; Hensley, S.E.; Nelson, M.I.; et al. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio 2019, 10, e00204–e00219. [Google Scholar] [CrossRef] [Green Version]
- Cruz, E.; Cain, J.; Crossett, B.; Kayser, V. Site-specific glycosylation profile of influenza A (H1N1) hemagglutinin through tandem mass spectrometry. Hum. Vaccin Immunother. 2018, 14, 508–517. [Google Scholar] [CrossRef]
- Thompson, A.J.; Cao, L.; Ma, Y.; Wang, X.; Diedrich, J.K.; Kikuchi, C.; Willis, S.; Worth, C.; McBride, R.; Yates, J.R., 3rd; et al. Human influenza virus memagglutinins contain conserved oligomannose N-linked glycans allowing potent neutralization by lectins. Cell Host Microbe 2020, 27, 725–735.e5. [Google Scholar] [CrossRef]
- Popova, L.; Smith, K.; West, A.H.; Wilson, P.C.; James, J.A.; Thompson, L.F.; Air, G.M. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE 2012, 7, e41895. [Google Scholar] [CrossRef]
- Chambers, B.S.; Parkhouse, K.; Ross, T.M.; Alby, K.; Hensley, S.E. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. 2015, 12, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashyap, A.K.; Steel, J.; Oner, A.F.; Dillon, M.A.; Swale, R.E.; Wall, K.M.; Perry, K.J.; Faynboym, A.; Ilhan, M.; Horowitz, M.; et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA 2008, 105, 5986–5991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [Green Version]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Lang, S.; Xie, J.; Zhu, X.; Wu, N.C.; Lerner, R.A.; Wilson, I.A. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep. 2017, 20, 2935–2943. [Google Scholar] [CrossRef] [Green Version]
- Pappas, L.; Foglierini, M.; Piccoli, L.; Kallewaard, N.L.; Turrini, F.; Silacci, C.; Fernandez-Rodriguez, B.; Agatic, G.; Giacchetto-Sasselli, I.; Pellicciotta, G.; et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014, 516, 418–422. [Google Scholar] [CrossRef]
- Avnir, Y.; Tallarico, A.S.; Zhu, Q.; Bennett, A.S.; Connelly, G.; Sheehan, J.; Sui, J.; Fahmy, A.; Huang, C.Y.; Cadwell, G.; et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 2014, 10, e1004103. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.C.; Yamayoshi, S.; Ito, M.; Uraki, R.; Kawaoka, Y.; Wilson, I.A. Recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus are encoded on the D3-9 segment of the Ig gene. Cell Host Microbe 2018, 24, 569–578.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesen, R.H.; Lee, P.S.; Stoop, E.J.; Hoffman, R.M.; Ekiert, D.C.; Bhabha, G.; Yu, W.; Juraszek, J.; Koudstaal, W.; Jongeneelen, M.; et al. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. USA 2014, 111, 445–450. [Google Scholar] [CrossRef] [Green Version]
- Joyce, M.G.; Wheatley, A.K.; Thomas, P.V.; Chuang, G.Y.; Soto, C.; Bailer, R.T.; Druz, A.; Georgiev, I.S.; Gillespie, R.A.; Kanekiyo, M.; et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 2016, 166, 609–623. [Google Scholar] [CrossRef] [Green Version]
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Neu, U.; McAuliffe, J.M.; Benjamin, E.; Wachter-Rosati, L.; Palmer-Hill, F.J.; Yuan, A.Q.; Walker, P.A.; et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 2016, 166, 596–608. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.F.; Joyce, M.G.; Chambers, M.J.; Gillespie, R.A.; Kanekiyo, M.; Leung, K.; Yang, E.S.; Tsybovsky, Y.; Wheatley, A.K.; Crank, M.C.; et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci. Immunol. 2017, 2, eaan2676. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.C.; Andrews, S.F.; Raab, J.E.; O’Connell, S.; Schramm, C.A.; Ding, X.; Chambers, M.J.; Leung, K.; Wang, L.; Zhang, Y.; et al. Convergent evolution in breadth of two VH6-1-encoded influenza antibody clonotypes from a single donor. Cell Host Microbe 2020, 28, 434–444.e4. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O’Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef]
- Lee, P.S.; Ohshima, N.; Stanfield, R.L.; Yu, W.; Iba, Y.; Okuno, Y.; Kurosawa, Y.; Wilson, I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014, 5, 3614. [Google Scholar] [CrossRef] [Green Version]
- Ohshima, N.; Iba, Y.; Kubota-Koketsu, R.; Asano, Y.; Okuno, Y.; Kurosawa, Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 2011, 85, 11048–11057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, K.R.; Watanabe, A.; Kuraoka, M.; Do, K.T.; McGee, C.E.; Sempowski, G.D.; Kepler, T.B.; Schmidt, A.G.; Kelsoe, G.; Harrison, S.C. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 2018, 48, 174–184.e9. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Yoshida, R.; Ekiert, D.C.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 2012, 109, 17040–17045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, R.; Igarashi, M.; Ozaki, H.; Kishida, N.; Tomabechi, D.; Kida, H.; Ito, K.; Takada, A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009, 5, e1000350. [Google Scholar] [CrossRef] [Green Version]
- Zost, S.J.; Lee, J.; Gumina, M.E.; Parkhouse, K.; Henry, C.; Wu, N.C.; Lee, C.D.; Wilson, I.A.; Wilson, P.C.; Bloom, J.D.; et al. Identification of antibodies targeting the H3N2 hemagglutinin receptor binding site following vaccination of humans. Cell Rep. 2019, 29, 4460–4470.e8. [Google Scholar] [CrossRef] [Green Version]
- Hong, M.; Lee, P.S.; Hoffman, R.M.; Zhu, X.; Krause, J.C.; Laursen, N.S.; Yoon, S.I.; Song, L.; Tussey, L.; Crowe, J.E., Jr.; et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 2013, 87, 12471–12480. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Basler, C.F.; Crowe, J.E., Jr. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 2011, 85, 10905–10908. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Albrecht, R.; Blum, D.L.; Ramos, I.; Fernandez-Sesma, A.; Edwards, K.M.; Garcia-Sastre, A.; et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J. Virol. 2012, 86, 6334–6340. [Google Scholar] [CrossRef] [Green Version]
- Whittle, J.R.; Zhang, R.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 2011, 108, 14216–14221. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.G.; Xu, H.; Khan, A.R.; O’Donnell, T.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Suphaphiphat, P.; Carfi, A.; et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. USA 2013, 110, 264–269. [Google Scholar] [CrossRef] [Green Version]
- Thornburg, N.J.; Zhang, H.; Bangaru, S.; Sapparapu, G.; Kose, N.; Lampley, R.M.; Bombardi, R.G.; Yu, Y.; Graham, S.; Branchizio, A.; et al. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J. Clin. Investig. 2016, 126, 1482–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, Y.; Liang, Y.; Ni, B.; Wan, Y.; Liao, Z.; Chan, K.H.; Yuen, K.Y.; Fu, X.; Shang, X.; et al. Fine antigenic variation within H5N1 influenza virus hemagglutinin’s antigenic sites defined by yeast cell surface display. Eur. J. Immunol. 2009, 39, 3498–3510. [Google Scholar] [CrossRef] [PubMed]
- Iba, Y.; Fujii, Y.; Ohshima, N.; Sumida, T.; Kubota-Koketsu, R.; Ikeda, M.; Wakiyama, M.; Shirouzu, M.; Okada, J.; Okuno, Y.; et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J. Virol. 2014, 88, 7130–7144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond, D.D.; Bajic, G.; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 2018, 115, 168–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, H.L.; Pallesen, J.; Lang, S.; Bangaru, S.; Urata, S.; Li, S.; Cottrell, C.A.; Bowman, C.A.; Crowe, J.E., Jr.; Wilson, I.A.; et al. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol. 2019, 17, e3000139. [Google Scholar] [CrossRef] [Green Version]
- Bangaru, S.; Lang, S.; Schotsaert, M.; Vanderven, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 2019, 177, 1136–1152.e8. [Google Scholar] [CrossRef]
- Watanabe, A.; McCarthy, K.R.; Kuraoka, M.; Schmidt, A.G.; Adachi, Y.; Onodera, T.; Tonouchi, K.; Caradonna, T.M.; Bajic, G.; Song, S.; et al. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 2019, 177, 1124–1135.e16. [Google Scholar] [CrossRef]
- Bajic, G.; Maron, M.J.; Adachi, Y.; Onodera, T.; McCarthy, K.R.; McGee, C.E.; Sempowski, G.D.; Takahashi, Y.; Kelsoe, G.; Kuraoka, M.; et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 2019, 25, 827–835.e6. [Google Scholar] [CrossRef]
- Das, D.K.; Govindan, R.; Nikic-Spiegel, I.; Krammer, F.; Lemke, E.A.; Munro, J.B. Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell 2018, 174, 926–937.e12. [Google Scholar] [CrossRef] [Green Version]
- Bodian, D.L.; Yamasaki, R.B.; Buswell, R.L.; Stearns, J.F.; White, J.M.; Kuntz, I.D. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry 1993, 32, 2967–2978. [Google Scholar] [CrossRef]
- Russell, R.J.; Kerry, P.S.; Stevens, D.J.; Steinhauer, D.A.; Martin, S.R.; Gamblin, S.J.; Skehel, J.J. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. USA 2008, 105, 17736–17741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagarinova, V.M.; Ignat’eva, G.S.; Sinitskaia, L.V.; Ivanova, A.M.; Rodina, M.A.; Tur’eva, A.V. [The new chemical preparation arbidol: Its prophylactic efficacy during influenza epidemics]. Zh Mikrobiol. Epidemiol. Immunobiol. 1993, 5, 40–43. [Google Scholar]
- Leneva, I.A.; Russell, R.J.; Boriskin, Y.S.; Hay, A.J. Characteristics of arbidol-resistant mutants of influenza virus: Implications for the mechanism of anti-influenza action of arbidol. Antivir. Res. 2009, 81, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Brancato, V.; Peduto, A.; Wharton, S.; Martin, S.; More, V.; Di Mola, A.; Massa, A.; Perfetto, B.; Donnarumma, G.; Schiraldi, C.; et al. Design of inhibitors of influenza virus membrane fusion: Synthesis, structure-activity relationship and in vitro antiviral activity of a novel indole series. Antivir. Res. 2013, 99, 125–135. [Google Scholar] [CrossRef]
- Kadam, R.U.; Wilson, I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA 2017, 114, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Wright, Z.V.F.; Wu, N.C.; Kadam, R.U.; Wilson, I.A.; Wolan, D.W. Structure-based optimization and synthesis of antiviral drug Arbidol analogues with significantly improved affinity to influenza hemagglutinin. Bioorg. Med. Chem. Lett. 2017, 27, 3744–3748. [Google Scholar] [CrossRef] [Green Version]
- Fleishman, S.J.; Whitehead, T.A.; Ekiert, D.C.; Dreyfus, C.; Corn, J.E.; Strauch, E.M.; Wilson, I.A.; Baker, D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 2011, 332, 816–821. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, T.A.; Chevalier, A.; Song, Y.; Dreyfus, C.; Fleishman, S.J.; De Mattos, C.; Myers, C.A.; Kamisetty, H.; Blair, P.; Wilson, I.A.; et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 2012, 30, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, A.; Silva, D.A.; Rocklin, G.J.; Hicks, D.R.; Vergara, R.; Murapa, P.; Bernard, S.M.; Zhang, L.; Lam, K.H.; Yao, G.; et al. Massively parallel de novo protein design for targeted therapeutics. Nature 2017, 550, 74–79. [Google Scholar] [CrossRef]
- Strauch, E.M.; Bernard, S.M.; La, D.; Bohn, A.J.; Lee, P.S.; Anderson, C.E.; Nieusma, T.; Holstein, C.A.; Garcia, N.K.; Hooper, K.A.; et al. Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat. Biotechnol. 2017, 35, 667–671. [Google Scholar] [CrossRef] [Green Version]
- Kadam, R.U.; Juraszek, J.; Brandenburg, B.; Buyck, C.; Schepens, W.B.G.; Kesteleyn, B.; Stoops, B.; Vreeken, R.J.; Vermond, J.; Goutier, W.; et al. Potent peptidic fusion inhibitors of influenza virus. Science 2017, 358, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dongen, M.J.P.; Kadam, R.U.; Juraszek, J.; Lawson, E.; Brandenburg, B.; Schmitz, F.; Schepens, W.B.G.; Stoops, B.; van Diepen, H.A.; Jongeneelen, M.; et al. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 2019, 363, eaar6221. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Kadam, R.U.; Lee, C.D.; Woehl, J.L.; Wu, N.C.; Zhu, X.; Kitamura, S.; Wilson, I.A.; Wolan, D.W. An influenza A hemagglutinin small-molecule fusion inhibitor identified by a new high-throughput fluorescence polarization screen. Proc. Natl. Acad. Sci. USA 2020, 117, 18431–18438. [Google Scholar] [CrossRef] [PubMed]
- Erbelding, E.J.; Post, D.; Stemmy, E.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef] [Green Version]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Valkenburg, S.A.; Mallajosyula, V.V.; Li, O.T.; Chin, A.W.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016, 6, 22666. [Google Scholar] [CrossRef] [Green Version]
- Corbett, K.S.; Moin, S.M.; Yassine, H.M.; Cagigi, A.; Kanekiyo, M.; Boyoglu-Barnum, S.; Myers, S.I.; Tsybovsky, Y.; Wheatley, A.K.; Schramm, C.A.; et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio 2019, 10, e02810–e02818. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.C.; Thompson, A.J.; Lee, J.M.; Su, W.; Arlian, B.M.; Xie, J.; Lerner, R.A.; Yen, H.L.; Bloom, J.D.; Wilson, I.A. Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science 2020, 368, 1335–1340. [Google Scholar] [CrossRef]
- Chai, N.; Swem, L.R.; Reichelt, M.; Chen-Harris, H.; Luis, E.; Park, S.; Fouts, A.; Lupardus, P.; Wu, T.D.; Li, O.; et al. Two escape mechanisms of influenza A virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog. 2016, 12, e1005702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.C.; Xie, J.; Zheng, T.; Nycholat, C.M.; Grande, G.; Paulson, J.C.; Lerner, R.A.; Wilson, I.A. Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe 2017, 21, 742–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doud, M.B.; Lee, J.M.; Bloom, J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018, 9, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeffe, J.R.; Van Rompay, K.K.A.; Olsen, P.C.; Wang, Q.; Gazumyan, A.; Azzopardi, S.A.; Schaefer-Babajew, D.; Lee, Y.E.; Stuart, J.B.; Singapuri, A.; et al. A combination of two human monoclonal antibodies prevents Zika virus escape mutations in non-human primates. Cell Rep. 2018, 25, 1385–1394.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [PubMed]
- Wang, Q.; Michailidis, E.; Yu, Y.; Wang, Z.; Hurley, A.M.; Oren, D.A.; Mayer, C.T.; Gazumyan, A.; Liu, Z.; Zhou, Y.; et al. A combination of human broadly neutralizing antibodies against hepatitis B virus HBsAg with distinct epitopes suppresses escape mutations. Cell Host Microbe 2020, 28, 335–349.e6. [Google Scholar] [CrossRef] [PubMed]
- Gilchuk, P.; Murin, C.D.; Milligan, J.C.; Cross, R.W.; Mire, C.E.; Ilinykh, P.A.; Huang, K.; Kuzmina, N.; Altman, P.X.; Hui, S.; et al. Analysis of a therapeutic antibody cocktail reveals determinants for cooperative and broad ebolavirus neutralization. Immunity 2020, 52, 388–403.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laursen, N.S.; Friesen, R.H.E.; Zhu, X.; Jongeneelen, M.; Blokland, S.; Vermond, J.; van Eijgen, A.; Tang, C.; van Diepen, H.; Obmolova, G.; et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 2018, 362, 598–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyoglu-Barnum, S.; Ellis, D.; Gillespie, R.A.; Hutchinson, G.B.; Park, Y.-J.; Moin, S.M.; Acton, O.; Ravichandran, R.; Murphy, M.; Pettie, D.; et al. Elicitation of broadly protective immunity to influenza by multivalent hemagglutinin nanoparticle vaccines. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jefferson, T.; Jones, M.A.; Doshi, P.; Del Mar, C.B.; Hama, R.; Thompson, M.J.; Spencer, E.A.; Onakpoya, I.; Mahtani, K.R.; Nunan, D.; et al. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst. Rev. 2014, 1, CD008965. [Google Scholar] [CrossRef] [Green Version]
- Webster, R.G.; Laver, W.G.; Kilbourne, E.D. Reactions of antibodies with surface antigens of influenza virus. J. Gen. Virol. 1968, 3, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Laver, W.G.; Colman, P.M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature 1983, 303, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Colman, P.M.; Varghese, J.N.; Laver, W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983, 303, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Colman, P.M.; Laver, W.G.; Varghese, J.N.; Baker, A.T.; Tulloch, P.A.; Air, G.M.; Webster, R.G. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 1987, 326, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, D.; Zhu, X.; McMahon, M.; Turner, J.S.; Wohlbold, T.J.; Schmitz, A.J.; Strohmeier, S.; Yu, W.; Nachbagauer, R.; Mudd, P.A.; et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 2019, 366, 499–504. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, N.C.; Wilson, I.A. Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure. Viruses 2020, 12, 1053. https://0-doi-org.brum.beds.ac.uk/10.3390/v12091053
Wu NC, Wilson IA. Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure. Viruses. 2020; 12(9):1053. https://0-doi-org.brum.beds.ac.uk/10.3390/v12091053
Chicago/Turabian StyleWu, Nicholas C., and Ian A. Wilson. 2020. "Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure" Viruses 12, no. 9: 1053. https://0-doi-org.brum.beds.ac.uk/10.3390/v12091053




